 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Kalax.0759s0011.2.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Saxifragales; Crassulaceae; Kalanchoe
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 531aa MW: 58119.5 Da PI: 8.118 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 51.3 | 2.1e-16 | 337 | 383 | 4 | 55 |
HHHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 4 ahnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
hn ErrRRdriN+++ L+el+P++ K +Ka++L +A+eY+ksLq
Kalax.0759s0011.2.p 337 VHNLSERRRRDRINEKMKALQELIPHS-----NKSDKASMLDEAIEYLKSLQ 383
6*************************7.....59*****************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF47459 | 3.01E-20 | 330 | 400 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene3D | G3DSA:4.10.280.10 | 8.6E-20 | 331 | 392 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS50888 | 17.942 | 333 | 382 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 1.58E-17 | 336 | 387 | No hit | No description |
| Pfam | PF00010 | 7.9E-14 | 337 | 383 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 1.2E-17 | 339 | 388 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009704 | Biological Process | de-etiolation | ||||
| GO:0010161 | Biological Process | red light signaling pathway | ||||
| GO:0010244 | Biological Process | response to low fluence blue light stimulus by blue low-fluence system | ||||
| GO:0010600 | Biological Process | regulation of auxin biosynthetic process | ||||
| GO:0010928 | Biological Process | regulation of auxin mediated signaling pathway | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 531 aa Download sequence Send to blast |
MNHCIPDWNF EDCLPAANQM KHIGPDHELM ELLWRNGQVV MHSQTHRKSG STAPNDPMAA 60 QKHDQSSFKG CESCWNPSNL IQDDETVSWL QYPLDDPLEK ESGSNLFIEL ADQVEANKHV 120 KQFENDNIYS KLNPFEANNG MPSVHQQPVV KHSAGPAMAT IPMAPRKIPM LSVPSSNPST 180 LNLTQHVNAG SSEVHFVAKR TDNIAHGGIR EGSTMTVGSS HCGSNLIANE VSRTSRNGVS 240 NKNISPTVFK NSSDRKNFPP KAKGQIDSPD PTMSSSSGGS GSSFMLSAGT TTNSHKRKAS 300 NIEDYECQSD VAELESASAK KQSQRSGSSR KTRAAEVHNL SERRRRDRIN EKMKALQELI 360 PHSNKSDKAS MLDEAIEYLK SLQLQLQMMW MSSGMQPMMY PAVQHYMSRM GIGMGPTMPS 420 MHNAMQLPRV PLLEQPMSGT TTPNQVNVCP TPVLTPLRYP NQLQNSTLAE QYARYLGFQQ 480 MQAASQPVNM FNFGGAQAFP SNHTIPLPGA GNAPPIAGAT MPDSLGGKLR * |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 341 | 346 | ERRRRD |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00606 | ChIP-seq | Transfer from AT2G43010 | Download |
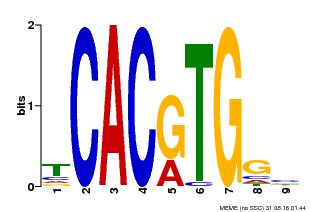
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_024184315.1 | 1e-161 | transcription factor PIF4-like | ||||
| Refseq | XP_024184316.1 | 1e-161 | transcription factor PIF4-like | ||||
| Refseq | XP_024184317.1 | 1e-161 | transcription factor PIF4-like | ||||
| TrEMBL | A0A2P6S0J7 | 1e-159 | A0A2P6S0J7_ROSCH; Putative transcription factor bHLH family | ||||
| STRING | XP_004303901.1 | 1e-152 | (Fragaria vesca) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G43010.2 | 8e-49 | phytochrome interacting factor 4 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Kalax.0759s0011.2.p |



