 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Kalax.0790s0005.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Saxifragales; Crassulaceae; Kalanchoe
|
||||||||
| Family | bZIP | ||||||||
| Protein Properties | Length: 395aa MW: 42151 Da PI: 7.9914 | ||||||||
| Description | bZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | bZIP_1 | 67.6 | 2e-21 | 250 | 312 | 1 | 63 |
XXXXCHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHH CS
bZIP_1 1 ekelkrerrkqkNReAArrsRqRKkaeieeLeekvkeLeaeNkaLkkeleelkkevaklksev 63
e+ lk errkq+NRe+ArrsR+RK+ae+eeL+ kv++L aeN +Lk+ +++l k+++klk e+
Kalax.0790s0005.1.p 250 ERHLKQERRKQSNRESARRSRLRKQAETEELAAKVDVLMAENVTLKSDISQLVKNSEKLKLEN 312
789*********************************************************987 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF07777 | 2.0E-29 | 1 | 96 | IPR012900 | G-box binding protein, multifunctional mosaic region |
| Gene3D | G3DSA:1.20.5.170 | 1.1E-13 | 248 | 309 | No hit | No description |
| Pfam | PF00170 | 6.8E-20 | 250 | 312 | IPR004827 | Basic-leucine zipper domain |
| SMART | SM00338 | 7.3E-17 | 250 | 314 | IPR004827 | Basic-leucine zipper domain |
| PROSITE profile | PS50217 | 12.128 | 252 | 315 | IPR004827 | Basic-leucine zipper domain |
| SuperFamily | SSF57959 | 2.63E-10 | 253 | 309 | No hit | No description |
| CDD | cd14702 | 1.57E-18 | 255 | 301 | No hit | No description |
| PROSITE pattern | PS00036 | 0 | 257 | 272 | IPR004827 | Basic-leucine zipper domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0005829 | Cellular Component | cytosol | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 395 aa Download sequence Send to blast |
MGNNEESKRC KSDKSTSPAP QDLIGMHVHP PADWAAMQAF YGPRIPLPQY YNSAYPAGHP 60 PHPYMWGLPQ AMMPSFAAPY AAIYPPGGVY AHPGVPIGSP KLGLGALSTP SATDALAATA 120 LSIQTPGKTS GNTTGGSMDS ESGDCGGSSC GSGVKTSGDE QSLIKRKGER PRGIEKTEKL 180 EIQANSTVAV DTNVVPSMVA GVGVGPASNG KLENVTALDM KTNHVVNSNL SVDAQQSGSK 240 LSSETCFQSE RHLKQERRKQ SNRESARRSR LRKQAETEEL AAKVDVLMAE NVTLKSDISQ 300 LVKNSEKLKL ENARMMEKLK NAKFEKAERI GPNDTDDNRN QTRSTENLLS RVNNSGSAVT 360 QEEEGEMYSN NNSNPAKLHQ LMDASPRADA VAAS* |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 266 | 272 | RRSRLRK |
| 2 | 266 | 273 | RRSRLRKQ |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00318 | DAP | Transfer from AT2G46270 | Download |
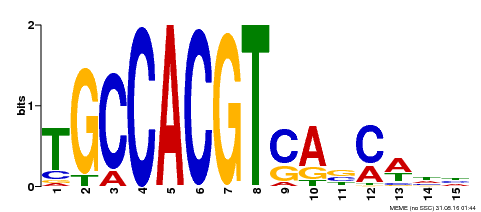
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_018827913.1 | 1e-112 | PREDICTED: common plant regulatory factor 1-like isoform X2 | ||||
| TrEMBL | A0A2I4F8E0 | 1e-111 | A0A2I4F8E0_JUGRE; common plant regulatory factor 1-like isoform X2 | ||||
| STRING | VIT_15s0046g01440.t01 | 1e-110 | (Vitis vinifera) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G46270.2 | 3e-52 | G-box binding factor 3 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Kalax.0790s0005.1.p |



