 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Kalax.0938s0013.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Saxifragales; Crassulaceae; Kalanchoe
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 222aa MW: 24760.4 Da PI: 4.6363 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 31.4 | 3.4e-10 | 53 | 96 | 7 | 55 |
HHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 7 erErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
Er+RR+r+ +++ Lr+l+P+ + ++Ka+i Av YI +Lq
Kalax.0938s0013.1.p 53 LTERNRRQRLSERLLVLRSLVPNI-----TNMNKATIIVDAVSYIEELQ 96
57*********************6.....58*****************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50888 | 14.373 | 46 | 95 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene3D | G3DSA:4.10.280.10 | 1.1E-14 | 46 | 113 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SuperFamily | SSF47459 | 2.22E-15 | 47 | 111 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 6.16E-10 | 48 | 96 | No hit | No description |
| SMART | SM00353 | 1.8E-10 | 52 | 101 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 1.1E-7 | 53 | 96 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0048658 | Biological Process | anther wall tapetum development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 222 aa Download sequence Send to blast |
MGMCGLAGQM KHKGVILYEP VKIGESSNEG KSASNEAVGE NGEVRRFKSK NLLTERNRRQ 60 RLSERLLVLR SLVPNITNMN KATIIVDAVS YIEELQGCVE ELTNQLQEME AKVCSDDEER 120 EFQEGETSLM QEIIQTPIQA EVDLTVINHS KLWMKLVFTN RRGGFTQLME ALNYLGLEPT 180 DVSLTTSGGA VLVSLCLEGI YGQDLEVNKI KELLQDIIRN I* |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00091 | SELEX | Transfer from AT4G21330 | Download |
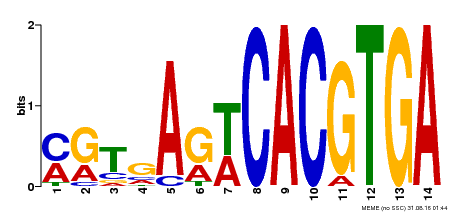
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_021809344.1 | 9e-54 | transcription factor DYT1-like | ||||
| TrEMBL | A0A2N9GEL9 | 3e-52 | A0A2N9GEL9_FAGSY; Uncharacterized protein | ||||
| STRING | XP_008225011.1 | 5e-53 | (Prunus mume) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G21330.1 | 3e-28 | bHLH family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Kalax.0938s0013.1.p |



