 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | LOC_Os01g14440.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza; Oryza sativa
|
||||||||
| Family | WRKY | ||||||||
| Protein Properties | Length: 584aa MW: 60502.4 Da PI: 8.7383 | ||||||||
| Description | WRKY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | WRKY | 97.1 | 1.2e-30 | 301 | 359 | 2 | 59 |
--SS-EEEEEEE--TT-SS-EEEEEE-ST.T---EEEEEE-SSSTTEEEEEEES--SS- CS
WRKY 2 dDgynWrKYGqKevkgsefprsYYrCtsa.gCpvkkkversaedpkvveitYegeHnhe 59
Dg++WrKYGqK+ kg+++pr+YYrCt+a gCpv+k+v+r+aed +++++tYeg+Hnh+
LOC_Os01g14440.1 301 ADGCQWRKYGQKMAKGNPCPRAYYRCTMAtGCPVRKQVQRCAEDRSILITTYEGTHNHP 359
7***************************99****************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:2.20.25.80 | 8.3E-34 | 285 | 361 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 5.62E-28 | 293 | 361 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 29.505 | 295 | 361 | IPR003657 | WRKY domain |
| SMART | SM00774 | 4.1E-37 | 300 | 360 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 3.7E-27 | 302 | 359 | IPR003657 | WRKY domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0010200 | Biological Process | response to chitin | ||||
| GO:0016036 | Biological Process | cellular response to phosphate starvation | ||||
| GO:0045892 | Biological Process | negative regulation of transcription, DNA-templated | ||||
| GO:0080169 | Biological Process | cellular response to boron-containing substance deprivation | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 584 aa Download sequence Send to blast |
MEADDSGGGG GRARRSVEVD FFSDEKKNMK KSRVSGGVAA EADDAKGPAA AGLAIKKEDL 60 TINVSRRRLL RSSIARHLFN SRRSNACARP CSLQLLPAGN NARSDRSMVV DDDAASRPDH 120 EEKSRSSNEL AAMQAELGRM NEENQRLRGM LTQVTTSYQA LQMHLVALMQ QRPQMMQPPT 180 QPEPPPPHQD GKAEGAVVPR QFLDLGPSSG AGGEAAEEPS NSSTEAGSPR RSSSTGNKDQ 240 ERGDSPDAPS TAAAWLPGRA MAPQMGAAGA AGKSHDQQAQ DANMRKARVS VRARSEAPII 300 ADGCQWRKYG QKMAKGNPCP RAYYRCTMAT GCPVRKQVQR CAEDRSILIT TYEGTHNHPL 360 PPAAMAMAST TSAAASMLLS GSMPSADGAA GLMSSNFLAR TVLPCSSSMA TISASAPFPT 420 VTLDLTHAPP GAPNAVPLNA ARPGAPAPQF QVPLPGGGMA PAFAVPPQVL YNQSKFSGLQ 480 MSSDSAEAAA AAAAAAQFAQ PRPPIGQLPG PLSDTVSAAA AAITADPNFT VALAAAITSI 540 IGGQHAAAAG NSNANNTNTN TTSNTNNTSS NNTTSNNTNS ETQ* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 2ayd_A | 1e-22 | 287 | 363 | 1 | 76 | WRKY transcription factor 1 |
| Search in ModeBase | ||||||
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Os.6144 | 0.0 | callus| leaf| panicle| root | ||||
| Expression -- Microarray ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | ID | E-value | ||||
| GEO | 50843961 | 0.0 | ||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00209 | DAP | Transfer from AT1G62300 | Download |
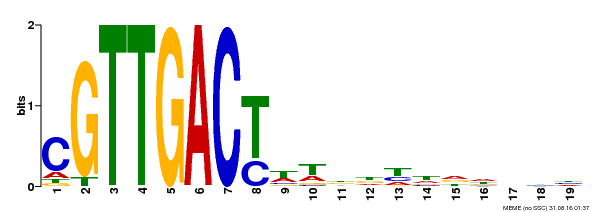
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | LOC_Os01g14440.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Phenotype -- Mutation ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | ID | |||||
| RiceGE | Os01g14440 | |||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AY676928 | 0.0 | AY676928.1 Oryza sativa (indica cultivar-group) transcription factor OsWRKY99 (OsWRKY99) mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015622141.1 | 0.0 | probable WRKY transcription factor 31 isoform X1 | ||||
| TrEMBL | Q0JP37 | 0.0 | Q0JP37_ORYSJ; Os01g0246700 protein | ||||
| STRING | ORGLA01G0083800.1 | 0.0 | (Oryza glaberrima) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP5265 | 36 | 56 | Representative plant | OGRP14 | 17 | 875 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G62300.1 | 6e-98 | WRKY family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | LOC_Os01g14440.1 |



