 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | LOC_Os02g35140.1 | ||||||||
| Common Name | ARF7, LOC4329672, Os02g0557200, OsJ_07141, OSJNBb0038F20.6 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza; Oryza sativa
|
||||||||
| Family | ARF | ||||||||
| Protein Properties | Length: 679aa MW: 74787.1 Da PI: 6.3855 | ||||||||
| Description | ARF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | B3 | 73.4 | 2.7e-23 | 128 | 228 | 1 | 98 |
EEEE-..-HHHHTT-EE--HHH.HTT.......---..--SEEEEEETTS-EEEEEE..EEETTEEEE-TTHHHHHHHHT--TT-EEEEEE-S CS
B3 1 ffkvltpsdvlksgrlvlpkkfaeeh.......ggkkeesktltledesgrsWevkliyrkksgryvltkGWkeFvkangLkegDfvvFkldg 86
f+k+lt sd++++g +++ +++aee+ ++++ + +l+ +d++g++W +++i+r++++r++lt+GW+ Fv++++L +gD ++F +
LOC_Os02g35140.1 128 FCKTLTASDTSTHGGFSVLRRHAEEClppldmtQNPPWQ--ELVARDLHGNEWHFRHIFRGQPRRHLLTTGWSVFVSSKRLVAGDAFIFL--R 216
99****************************986555545..8************************************************..4 PP
SSEE..EEEEE- CS
B3 87 rsefelvvkvfr 98
+++el+v+v+r
LOC_Os02g35140.1 217 GENGELRVGVRR 228
49999*****97 PP
| |||||||
| 2 | Auxin_resp | 111.4 | 9e-37 | 254 | 336 | 1 | 83 |
Auxin_resp 1 aahaastksvFevvYnPrastseFvvkvekvekalkvkvsvGmRfkmafetedsserrlsGtvvgvsdldpvrWpnSkWrsLk 83
a+ha+st++ F+v+Y+Pr+s+seFvv+++k+++a ++k+svGmRfkm+fe+++++err+sGt++gv ++++ W nS WrsLk
LOC_Os02g35140.1 254 ASHAISTGTLFSVFYKPRTSQSEFVVSANKYLEAKNSKISVGMRFKMRFEGDEAPERRFSGTIIGVGSMSTSPWANSDWRSLK 336
789******************************************************************************96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF101936 | 4.58E-44 | 117 | 257 | IPR015300 | DNA-binding pseudobarrel domain |
| Gene3D | G3DSA:2.40.330.10 | 4.6E-41 | 123 | 243 | IPR015300 | DNA-binding pseudobarrel domain |
| CDD | cd10017 | 7.75E-22 | 126 | 228 | No hit | No description |
| Pfam | PF02362 | 5.4E-21 | 128 | 228 | IPR003340 | B3 DNA binding domain |
| PROSITE profile | PS50863 | 11.462 | 128 | 230 | IPR003340 | B3 DNA binding domain |
| SMART | SM01019 | 1.5E-22 | 128 | 230 | IPR003340 | B3 DNA binding domain |
| Pfam | PF06507 | 1.9E-34 | 254 | 336 | IPR010525 | Auxin response factor |
| Pfam | PF02309 | 9.9E-13 | 535 | 639 | IPR033389 | AUX/IAA domain |
| PROSITE profile | PS51745 | 25.456 | 548 | 641 | IPR000270 | PB1 domain |
| SuperFamily | SSF54277 | 6.54E-12 | 550 | 627 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009734 | Biological Process | auxin-activated signaling pathway | ||||
| GO:0010150 | Biological Process | leaf senescence | ||||
| GO:0045892 | Biological Process | negative regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0042802 | Molecular Function | identical protein binding | ||||
| Plant Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PO Term | PO Category | PO Description | ||||
| PO:0009066 | anatomy | anther | ||||
| PO:0001004 | developmental stage | anther development stage | ||||
| PO:0007130 | developmental stage | sporophyte reproductive stage | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 679 aa Download sequence Send to blast |
MAGSVVAAAA AAGGGTGSSC DALYRELWHA CAGPLVTVPR QGELVYYFPQ GHMEQLEAST 60 DQQLDQHLPL FNLPSKILCK VVNVELRAET DSDEVYAQIM LQPEADQNEL TSPKPEPHEP 120 EKCNVHSFCK TLTASDTSTH GGFSVLRRHA EECLPPLDMT QNPPWQELVA RDLHGNEWHF 180 RHIFRGQPRR HLLTTGWSVF VSSKRLVAGD AFIFLRGENG ELRVGVRRLM RQLNNMPSSV 240 ISSHSMHLGV LATASHAIST GTLFSVFYKP RTSQSEFVVS ANKYLEAKNS KISVGMRFKM 300 RFEGDEAPER RFSGTIIGVG SMSTSPWANS DWRSLKVQWD EPSVVPRPDR VSPWELEPLA 360 VSNSQPSPQP PARNKRARPP ASNSIAPELP PVFGLWKSSA ESTQGFSFSG LQRTQELYPS 420 SPNPIFSTSL NVGFSTKNEP SALSNKHFYW PMRETRANSY SASISKVPSE KKQEPSSAGC 480 RLFGIEISSA VEATSPLAAV SGVGQDQPAA SVDAESDQLS QPSHANKSDA PAASSEPSPH 540 ETQSRQVRSC TKVIMQGMAV GRAVDLTRLH GYDDLRCKLE EMFDIQGELS ASLKKWKVVY 600 TDDEDDMMLV GDDPWPEFCS MVKRIYIYTY EEAKQLTPKS KLPIIGDAIK PNPNKQSPES 660 DMPHSDLDST APVTDKDC* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 4ldv_A | 0.0 | 5 | 359 | 1 | 355 | Auxin response factor 1 |
| 4ldw_A | 0.0 | 5 | 359 | 1 | 355 | Auxin response factor 1 |
| 4ldw_B | 0.0 | 5 | 359 | 1 | 355 | Auxin response factor 1 |
| Search in ModeBase | ||||||
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Os.59273 | 0.0 | callus| flower| leaf| panicle| root| seed| stem | ||||
| Expression -- Microarray ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | ID | E-value | ||||
| GEO | 115446714 | 0.0 | ||||
| Expression Atlas | Q6YVY0 | - | ||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | TISSUE SPECIFICITY: Expressed in roots, culms, leaves and young panicles. {ECO:0000269|PubMed:17408882}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Auxin response factors (ARFs) are transcriptional factors that bind specifically to the DNA sequence 5'-TGTCTC-3' found in the auxin-responsive promoter elements (AuxREs). | |||||
| Function -- GeneRIF ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
|
||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00097 | PBM | Transfer from AT1G59750 | Download |
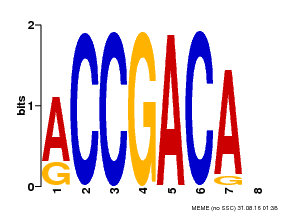
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | LOC_Os02g35140.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | - | ||||
| Phenotype -- Mutation ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | ID | |||||
| RiceGE | Os02g35140 | |||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK070026 | 0.0 | AK070026.1 Oryza sativa Japonica Group cDNA clone:J023036G16, full insert sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015625501.1 | 0.0 | auxin response factor 7 | ||||
| Swissprot | Q6YVY0 | 0.0 | ARFG_ORYSJ; Auxin response factor 7 | ||||
| TrEMBL | A0A0E0NGE4 | 0.0 | A0A0E0NGE4_ORYRU; Auxin response factor | ||||
| STRING | ORUFI02G21640.1 | 0.0 | (Oryza rufipogon) | ||||
| STRING | OS02T0557200-01 | 0.0 | (Oryza sativa) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP1082 | 36 | 116 | Representative plant | OGRP550 | 15 | 82 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G59750.4 | 0.0 | auxin response factor 1 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | LOC_Os02g35140.1 |
| Entrez Gene | 4329672 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



