 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | LOC_Os02g43820.1 | ||||||||
| Common Name | LOC4330191, OJ1003_B06.21, Os02g0655200, P0519E06.5 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza; Oryza sativa
|
||||||||
| Family | ERF | ||||||||
| Protein Properties | Length: 345aa MW: 36329.6 Da PI: 5.7499 | ||||||||
| Description | ERF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 64.3 | 2.5e-20 | 154 | 205 | 2 | 56 |
AP2 2 gykGVrwdkkrgrWvAeIrdpsengkrkrfslgkfgtaeeAakaaiaarkklege 56
+y+GVr+++ +g+++AeIrdp+++g r++lg+++ta eAa+a+++a+ +++g+
LOC_Os02g43820.1 154 KYRGVRQRP-WGKFAAEIRDPKKRG--SRVWLGTYDTAIEAARAYDRAAFRMRGA 205
8********.***********8876..**************************95 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:3.30.730.10 | 1.4E-32 | 153 | 213 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 8.5E-38 | 154 | 218 | IPR001471 | AP2/ERF domain |
| Pfam | PF00847 | 1.6E-14 | 154 | 204 | IPR001471 | AP2/ERF domain |
| SuperFamily | SSF54171 | 2.42E-22 | 154 | 213 | IPR016177 | DNA-binding domain |
| PROSITE profile | PS51032 | 24.25 | 154 | 212 | IPR001471 | AP2/ERF domain |
| CDD | cd00018 | 1.90E-31 | 154 | 213 | No hit | No description |
| PRINTS | PR00367 | 2.1E-13 | 155 | 166 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 2.1E-13 | 194 | 214 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009409 | Biological Process | response to cold | ||||
| GO:0010200 | Biological Process | response to chitin | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Plant Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PO Term | PO Category | PO Description | ||||
| PO:0009089 | anatomy | endosperm | ||||
| PO:0007042 | developmental stage | whole plant fruit formation stage | ||||
| PO:0007633 | developmental stage | endosperm development stage | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 345 aa Download sequence Send to blast |
MDFSGDIDDL ISLQLLHDQL LGVEADACLP VAVHHDGVAA FAEHQQGFHP AAFLPQQPMT 60 MTPAGYVDMA NDQYLGAHAA AGEAEAVYRA AAAEPVMIRF GGEVSPVSDP RRPPLTISLP 120 PTSHAWAAAE AVHPAALLQA QTAAAAADPN DFRKYRGVRQ RPWGKFAAEI RDPKKRGSRV 180 WLGTYDTAIE AARAYDRAAF RMRGAKAILN FPNEVGSRGA DFLAPPPPPP PPTTSTHGKR 240 KRHETAAADP DVEVIGESSK SVKTETYTSP ASSSLASTTT STVTSSSTSP SPSSEAAACG 300 GGGGELFVPP MPSSWSWDQL EGFFGILSPL SPHPQMGFPE VAVN* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 2gcc_A | 1e-27 | 155 | 217 | 6 | 68 | ATERF1 |
| 3gcc_A | 1e-27 | 155 | 217 | 6 | 68 | ATERF1 |
| Search in ModeBase | ||||||
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Os.38982 | 0.0 | callus| panicle | ||||
| Expression -- Microarray ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | ID | E-value | ||||
| GEO | 33440023 | 0.0 | ||||
| Expression Atlas | Q6H6I3 | - | ||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00549 | DAP | Transfer from AT5G47230 | Download |
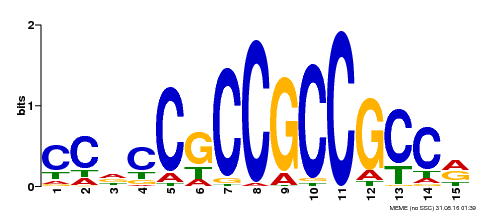
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | LOC_Os02g43820.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Phenotype -- Mutation ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | ID | |||||
| RiceGE | Os02g43820 | |||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AP004676 | 0.0 | AP004676.3 Oryza sativa Japonica Group genomic DNA, chromosome 2, BAC clone:OJ1003_B06. | |||
| GenBank | AP005006 | 0.0 | AP005006.3 Oryza sativa Japonica Group genomic DNA, chromosome 2, PAC clone:P0519E06. | |||
| GenBank | AP014958 | 0.0 | AP014958.1 Oryza sativa Japonica Group DNA, chromosome 2, cultivar: Nipponbare, complete sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015624058.1 | 0.0 | ethylene-responsive transcription factor 5 | ||||
| TrEMBL | Q6H6I3 | 0.0 | Q6H6I3_ORYSJ; Os02g0655200 protein | ||||
| STRING | ORUFI02G27860.1 | 0.0 | (Oryza rufipogon) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP103 | 37 | 465 | Representative plant | OGRP6 | 16 | 1718 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G47230.1 | 1e-24 | ethylene responsive element binding factor 5 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | LOC_Os02g43820.1 |
| Entrez Gene | 4330191 |



