 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | LOC_Os04g43680.1 | ||||||||
| Common Name | LOC4336407, LTR1, MYB4, Os04g0517100, OSJNBa0073E02.6 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza; Oryza sativa
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 258aa MW: 27914.4 Da PI: 7.0559 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 54.9 | 2e-17 | 14 | 61 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g+WT+eEd+ lv ++++G g+W++ ++ g+ R++k+c++rw +yl
LOC_Os04g43680.1 14 KGPWTPEEDKVLVAHIQRHGHGNWRALPKQAGLLRCGKSCRLRWINYL 61
79******************************99************97 PP
| |||||||
| 2 | Myb_DNA-binding | 51.3 | 2.6e-16 | 67 | 112 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
rg++++eE++ +++++++lG++ W++Ia++++ gRt++++k+ w+++l
LOC_Os04g43680.1 67 RGNFSKEEEDTIIHLHELLGNR-WSAIAARLP-GRTDNEIKNVWHTHL 112
89********************.*********.************986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.60 | 6.3E-26 | 5 | 64 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 16.37 | 9 | 61 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 3.23E-31 | 10 | 108 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 2.3E-13 | 13 | 63 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 1.6E-15 | 14 | 61 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 5.34E-11 | 16 | 61 | No hit | No description |
| PROSITE profile | PS51294 | 25.291 | 62 | 116 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 2.6E-26 | 65 | 116 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 1.1E-14 | 66 | 114 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 2.0E-15 | 67 | 112 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 5.57E-11 | 69 | 112 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0009733 | Biological Process | response to auxin | ||||
| GO:0009753 | Biological Process | response to jasmonic acid | ||||
| GO:0010200 | Biological Process | response to chitin | ||||
| GO:0046686 | Biological Process | response to cadmium ion | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Plant Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PO Term | PO Category | PO Description | ||||
| PO:0009037 | anatomy | lemma | ||||
| PO:0001047 | developmental stage | lemma development stage | ||||
| PO:0007130 | developmental stage | sporophyte reproductive stage | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 258 aa Download sequence Send to blast |
MGRAPCCEKM GLKKGPWTPE EDKVLVAHIQ RHGHGNWRAL PKQAGLLRCG KSCRLRWINY 60 LRPDIKRGNF SKEEEDTIIH LHELLGNRWS AIAARLPGRT DNEIKNVWHT HLKKRLDAPA 120 QGGHVAASGG KKHKKPKSAK KPAAAAAAPP ASPERSASSS VTESSMASSV AEEHGNAGIS 180 SASASVCAKE ESSFTSASEE FQIDDSFWSE TLSMPLDGYD VSMEPGDAFV APPSADDMDY 240 WLGVFMESGE AQDLPQI* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1a5j_A | 1e-26 | 12 | 118 | 5 | 110 | B-MYB |
| Search in ModeBase | ||||||
| Expression -- Microarray ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | ID | |||||
| Expression Atlas | Q7XBH4 | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional activator involved in cold stress response (PubMed:14675437, PubMed:20807373, PubMed:22246661). Regulates positively the expression of genes involved in reactive oxygen species (ROS) scavenging such as peroxidase and superoxide dismutase during cold stress. Transactivates a complex gene network that have major effects on stress tolerance and panicle development (PubMed:20807373). {ECO:0000269|PubMed:14675437, ECO:0000269|PubMed:20807373, ECO:0000269|PubMed:22246661}. | |||||
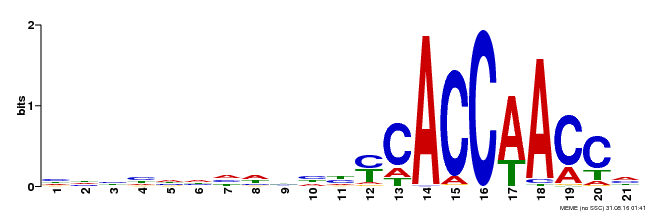
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00375 | DAP | Transfer from AT3G23250 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | LOC_Os04g43680.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By cold stress. {ECO:0000269|PubMed:14675437, ECO:0000269|PubMed:20807373, ECO:0000269|Ref.2}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Phenotype -- Mutation ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | ID | |||||
| RiceGE | Os04g43680 | |||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | Y11414 | 0.0 | Y11414.1 O.sativa mRNA for myb factor, 1202 bp. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015633465.1 | 0.0 | transcription factor MYB4 | ||||
| Swissprot | Q7XBH4 | 0.0 | MYB4_ORYSJ; Transcription factor MYB4 | ||||
| TrEMBL | Q01J53 | 0.0 | Q01J53_ORYSA; OSIGBa0145M07.4 protein | ||||
| STRING | ORGLA04G0158700.1 | 0.0 | (Oryza glaberrima) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP79 | 38 | 563 | Representative plant | OGRP5 | 17 | 1784 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G23250.1 | 3e-76 | myb domain protein 15 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | LOC_Os04g43680.1 |
| Entrez Gene | 4336407 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



