 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | LOC_Os05g36160.2 | ||||||||
| Common Name | OJ1058_F05.8, Os05g0437700, OSJNBb0042J17.2, OSNPB_050437700 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza; Oryza sativa
|
||||||||
| Family | bZIP | ||||||||
| Protein Properties | Length: 248aa MW: 25159.1 Da PI: 6.0935 | ||||||||
| Description | bZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | bZIP_1 | 45.2 | 2e-14 | 186 | 237 | 3 | 54 |
XXCHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHH CS
bZIP_1 3 elkrerrkqkNReAArrsRqRKkaeieeLeekvkeLeaeNkaLkkeleelkk 54
+ +r++r++kNRe+A rsR+RK+a+i+eLe v+eLe+e +L e ee ++
LOC_Os05g36160.2 186 AMQRQKRMIKNRESAARSRERKQAYIAELEAQVAELEEEHAQLLREQEEKNQ 237
689**************************************99977666444 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00338 | 2.8E-8 | 184 | 245 | IPR004827 | Basic-leucine zipper domain |
| PROSITE profile | PS50217 | 10.875 | 186 | 234 | IPR004827 | Basic-leucine zipper domain |
| Pfam | PF00170 | 2.3E-12 | 186 | 236 | IPR004827 | Basic-leucine zipper domain |
| SuperFamily | SSF57959 | 6.96E-11 | 188 | 237 | No hit | No description |
| CDD | cd14707 | 1.64E-17 | 188 | 234 | No hit | No description |
| Gene3D | G3DSA:1.20.5.170 | 2.6E-13 | 189 | 235 | No hit | No description |
| PROSITE pattern | PS00036 | 0 | 191 | 206 | IPR004827 | Basic-leucine zipper domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Plant Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PO Term | PO Category | PO Description | ||||
| PO:0009089 | anatomy | endosperm | ||||
| PO:0007042 | developmental stage | whole plant fruit formation stage | ||||
| PO:0007633 | developmental stage | endosperm development stage | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 248 aa Download sequence Send to blast |
MASSRVMAAA AASSSSSPPP PPPAAAAAGG AADLARFRST SSGIGSMNMD DILRNIYGEA 60 APPPGAAGSA PAPPPAGEAA GAPVAEVAAR RTAEEVWKEI SSSGGLSAPA PAPAAGAAGR 120 GGGPEMTLED FLAREDDPRA TAVEGNMVVG FPNVTEGVGT AGGGRGGGGG GRGRKRTLMD 180 PADRAAMQRQ KRMIKNRESA ARSRERKQAY IAELEAQVAE LEEEHAQLLR EQEEKNQKRL 240 KEVSLIV* |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 162 | 170 | GGRGGGGGG |
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Os.37925 | 0.0 | callus| leaf| panicle| seed | ||||
| Expression -- Microarray ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | ID | E-value | ||||
| GEO | 37990279 | 0.0 | ||||
| Expression Atlas | Q75HX9 | - | ||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator that binds to the ABA-responsive elements (ABREs) in vitro. Involved in abiotic stress responses and abscisic acid (ABA) signaling (PubMed:20039193). Involved in the signaling pathway that induces growth inhibition in response to D-allose (PubMed:23397192). {ECO:0000269|PubMed:20039193, ECO:0000269|PubMed:23397192}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00648 | PBM | 25215497 | Download |
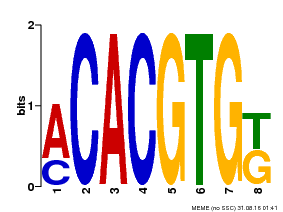
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | LOC_Os05g36160.2 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Induced by anoxia, drought, salt stress, oxidative stress, cold and abscisic acid (ABA) (PubMed:20039193). Induced by D-allose (PubMed:23397192). {ECO:0000269|PubMed:20039193, ECO:0000269|PubMed:23397192}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | - | ||||
| Phenotype -- Mutation ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | ID | |||||
| RiceGE | Os05g36160 | |||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK120656 | 0.0 | AK120656.1 Oryza sativa Japonica Group cDNA clone:J013158B06, full insert sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015639395.1 | 1e-167 | bZIP transcription factor 12-like | ||||
| Swissprot | Q0JHF1 | 5e-51 | BZP12_ORYSJ; bZIP transcription factor 12 | ||||
| TrEMBL | Q75HX9 | 1e-169 | Q75HX9_ORYSJ; Os05g0437700 protein | ||||
| STRING | OS05T0437700-01 | 1e-170 | (Oryza sativa) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G44080.1 | 5e-12 | bZIP family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | LOC_Os05g36160.2 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



