 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | LOC_Os07g39480.1 | ||||||||
| Common Name | LOC4343736, OJ1127_E01.114, Os07g0583700, OsJ_24901, OSNPB_070583700 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza; Oryza sativa
|
||||||||
| Family | WRKY | ||||||||
| Protein Properties | Length: 619aa MW: 66163.6 Da PI: 6.3317 | ||||||||
| Description | WRKY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | WRKY | 101.2 | 6.3e-32 | 236 | 293 | 2 | 60 |
--SS-EEEEEEE--TT-SS-EEEEEE-STT---EEEEEE-SSSTTEEEEEEES--SS-- CS
WRKY 2 dDgynWrKYGqKevkgsefprsYYrCtsagCpvkkkversaedpkvveitYegeHnhek 60
+DgynWrKYGqK+vkgse+prsYY+Ct+++C vkk +ers d++++e++Y+g+Hnh+k
LOC_Os07g39480.1 236 EDGYNWRKYGQKHVKGSENPRSYYKCTHPNCDVKKLLERSL-DGQITEVVYKGRHNHPK 293
8****************************************.***************85 PP
| |||||||
| 2 | WRKY | 106.4 | 1.5e-33 | 409 | 467 | 1 | 59 |
---SS-EEEEEEE--TT-SS-EEEEEE-STT---EEEEEE-SSSTTEEEEEEES--SS- CS
WRKY 1 ldDgynWrKYGqKevkgsefprsYYrCtsagCpvkkkversaedpkvveitYegeHnhe 59
ldDgy+WrKYGqK+vkg+++prsYY+Ct +gCpv+k+ver+++dpk v++tYeg+Hnhe
LOC_Os07g39480.1 409 LDDGYRWRKYGQKVVKGNPNPRSYYKCTNTGCPVRKHVERASHDPKSVITTYEGKHNHE 467
59********************************************************8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:2.20.25.80 | 1.2E-27 | 229 | 294 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 23.789 | 230 | 294 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 9.02E-25 | 234 | 294 | IPR003657 | WRKY domain |
| SMART | SM00774 | 8.3E-35 | 235 | 293 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 3.5E-24 | 236 | 292 | IPR003657 | WRKY domain |
| Gene3D | G3DSA:2.20.25.80 | 1.1E-37 | 394 | 469 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 1.44E-29 | 401 | 469 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 38.526 | 404 | 469 | IPR003657 | WRKY domain |
| SMART | SM00774 | 1.7E-39 | 409 | 468 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 2.5E-26 | 410 | 467 | IPR003657 | WRKY domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009611 | Biological Process | response to wounding | ||||
| GO:0009961 | Biological Process | response to 1-aminocyclopropane-1-carboxylic acid | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Plant Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PO Term | PO Category | PO Description | ||||
| PO:0009049 | anatomy | inflorescence | ||||
| PO:0001083 | developmental stage | inflorescence development stage | ||||
| PO:0007006 | developmental stage | IL.00 inflorescence just visible stage | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 619 aa Download sequence Send to blast |
MADSPNPSSG DHPAGVGGSP EKQPPVDRRV AALAAGAAGA GARYKAMSPA RLPISREPCL 60 TIPAGFSPSA LLESPVLLTN FKVEPSPTTG TLSMAAIMNK SANPDILPSP RDKTSGSTHE 120 DGGSRDFEFK PHLNSSSQST ASAINDPKKH ETSMKNESLN TALSSDDMMI DNIPLCSRES 180 TLAVNISSAP SQLVGMVGLT DSSPAEVGTS ELHQMNSSGN AMQESQPESV AEKSAEDGYN 240 WRKYGQKHVK GSENPRSYYK CTHPNCDVKK LLERSLDGQI TEVVYKGRHN HPKPQPNRRL 300 SAGAVPPIQG EERYDGVATT DDKSSNVLSI LGNAVHTAGM IEPVPGSASD DDNDAGGGRP 360 YPGDDAVEDD DLESKRRKME SAAIDAALMG KPNREPRVVV QTVSEVDILD DGYRWRKYGQ 420 KVVKGNPNPR SYYKCTNTGC PVRKHVERAS HDPKSVITTY EGKHNHEVPA SRNASHEMST 480 PPMKPVVHPI NSNMQGLGGM MRACEPRTFP NQYSQAAESD TISLDLGVGI SPNHSDATNQ 540 LQSSVSDQMQ YQMQPMGSVY SNMGLPAMAM PTMAGNAASN IYGSREEKPS EGFTFKATPM 600 DHSANLCYST AGNLVMGP* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wj2_A | 8e-37 | 236 | 470 | 8 | 78 | Probable WRKY transcription factor 4 |
| 2lex_A | 8e-37 | 236 | 470 | 8 | 78 | Probable WRKY transcription factor 4 |
| Search in ModeBase | ||||||
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Os.18862 | 0.0 | callus| flower| leaf| panicle| root| seed| stem | ||||
| Expression -- Microarray ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | ID | E-value | ||||
| GEO | 32980561 | 0.0 | ||||
| Expression Atlas | Q84ZS7 | - | ||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | DEVELOPMENTAL STAGE: Peak of expression 12 days after pollination. {ECO:0000269|PubMed:12953112}. | |||||
| Uniprot | TISSUE SPECIFICITY: Expressed in endosperm, but not in leaves. {ECO:0000269|PubMed:12953112}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor involved in starch synthesis (PubMed:12953112). Acts as a transcriptional activator in sugar signaling (PubMed:16167901). Interacts specifically with the SURE and W-box elements, but not with the SP8a element (PubMed:12953112). {ECO:0000269|PubMed:12953112, ECO:0000269|PubMed:16167901}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00455 | DAP | Transfer from AT4G26640 | Download |
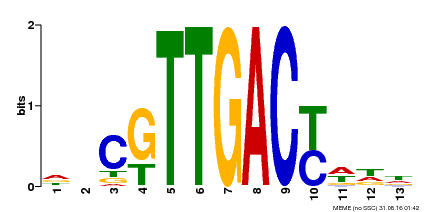
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | LOC_Os07g39480.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Up-regulated by sugar. {ECO:0000269|PubMed:12953112}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Phenotype -- Mutation ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | ID | |||||
| RiceGE | Os07g39480 | |||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK070537 | 0.0 | AK070537.1 Oryza sativa Japonica Group cDNA clone:J023062H03, full insert sequence. | |||
| GenBank | AY341850 | 0.0 | AY341850.1 Oryza sativa (japonica cultivar-group) WRKY9 mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015646742.1 | 0.0 | WRKY transcription factor SUSIBA2 isoform X1 | ||||
| Swissprot | Q6VWJ6 | 0.0 | WRK46_HORVU; WRKY transcription factor SUSIBA2 | ||||
| TrEMBL | Q5HZ67 | 0.0 | Q5HZ67_ORYSA; WRKY transcription factor 78 | ||||
| TrEMBL | Q84ZS7 | 0.0 | Q84ZS7_ORYSJ; Os07g0583700 protein | ||||
| STRING | OS07T0583700-01 | 0.0 | (Oryza sativa) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP6740 | 38 | 51 | Representative plant | OGRP14 | 17 | 875 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G26640.2 | 1e-125 | WRKY family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | LOC_Os07g39480.1 |
| Entrez Gene | 4343736 |



