 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | LOC_Os09g29930.3 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza; Oryza sativa
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 144aa MW: 16074 Da PI: 9.1399 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 58.5 | 1.2e-18 | 56 | 105 | 2 | 55 |
HHHHHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 2 rrahnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
r +h++ E+rRR++iN++f+ LrellP++ ++K +Ka+ L +++eYI+ Lq
LOC_Os09g29930.3 56 RSKHSATEQRRRSKINDRFQILRELLPHS----DQKRDKATFLLEVIEYIRFLQ 105
899*************************9....9*******************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF47459 | 3.53E-20 | 51 | 123 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene3D | G3DSA:4.10.280.10 | 3.7E-25 | 52 | 125 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 6.32E-14 | 52 | 109 | No hit | No description |
| PROSITE profile | PS50888 | 16.952 | 54 | 104 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 4.4E-16 | 56 | 105 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 2.6E-13 | 60 | 110 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006351 | Biological Process | transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 144 aa Download sequence Send to blast |
MDSGSRSSGG AGFDDDDGVA ARREVSSSLK ELTVRVDGKG GSCSGSGTDQ RPSSPRSKHS 60 ATEQRRRSKI NDRFQILREL LPHSDQKRDK ATFLLEVIEY IRFLQEKVQK FEASVPEWNQ 120 ENAKILPWVI TTHFYPPKPA LLD* |
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Os.16741 | 1e-165 | callus| flower| leaf| panicle| root| seed| stem | ||||
| Expression -- Microarray ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | ID | E-value | ||||
| GEO | 32987105 | 1e-165 | ||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | TISSUE SPECIFICITY: Expressed constitutively in roots. {ECO:0000269|PubMed:12679534}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Positive brassinosteroid-signaling protein. Transcription factor that bind specifically to the DNA sequence 5'-CANNTG-3'(E box). Can bind individually to the promoter as a homodimer or synergistically as a heterodimer with BZR2/BES1. Does not itself activate transcription but enhances BZR2/BES1-mediated target gene activation. {ECO:0000269|PubMed:15680330}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00219 | DAP | Transfer from AT1G69010 | Download |
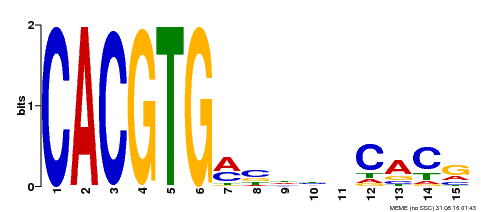
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | LOC_Os09g29930.3 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Repressed by cold treatment. {ECO:0000269|PubMed:12679534}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Phenotype -- Mutation ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | ID | |||||
| RiceGE | Os09g29930 | |||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK071315 | 0.0 | AK071315.1 Oryza sativa Japonica Group cDNA clone:J023084K14, full insert sequence. | |||
| GenBank | EU837261 | 0.0 | EU837261.1 Oryza sativa Japonica Group clone KCB846A08 bHLH transcription factor mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015612524.1 | 9e-86 | transcription factor BIM3 isoform X1 | ||||
| Refseq | XP_015612525.1 | 9e-87 | transcription factor BIM2 isoform X2 | ||||
| Swissprot | Q9LEZ3 | 2e-34 | BIM1_ARATH; Transcription factor BIM1 | ||||
| TrEMBL | B7EJ81 | 6e-87 | B7EJ81_ORYSJ; cDNA clone:J023084K14, full insert sequence | ||||
| STRING | ORUFI09G14830.1 | 1e-85 | (Oryza rufipogon) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G08130.3 | 2e-37 | bHLH family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | LOC_Os09g29930.3 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



