 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | LOC_Os11g47460.2 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza; Oryza sativa
|
||||||||
| Family | MYB_related | ||||||||
| Protein Properties | Length: 207aa MW: 23288.5 Da PI: 8.8025 | ||||||||
| Description | MYB_related family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 53.9 | 4.1e-17 | 33 | 77 | 2 | 48 |
SSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 2 grWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
gr+++ E+ l++++++++G++ W++Iar+++ gRt++++k++w++++
LOC_Os11g47460.2 33 GRMSPKEEHLIIELHARWGNR-WSRIARRLP-GRTDNEIKNYWRTHM 77
89*******************.*********.************986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 2.21E-21 | 12 | 82 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 2.9E-9 | 12 | 39 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50090 | 4.426 | 12 | 26 | IPR017877 | Myb-like domain |
| PROSITE profile | PS51294 | 25.581 | 27 | 81 | IPR017930 | Myb domain |
| SMART | SM00717 | 1.7E-14 | 31 | 79 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 1.6E-15 | 33 | 76 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 1.82E-11 | 34 | 77 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 4.2E-20 | 40 | 78 | IPR009057 | Homeodomain-like |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0009739 | Biological Process | response to gibberellin | ||||
| GO:0009751 | Biological Process | response to salicylic acid | ||||
| GO:0009753 | Biological Process | response to jasmonic acid | ||||
| GO:0010200 | Biological Process | response to chitin | ||||
| GO:0046686 | Biological Process | response to cadmium ion | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 207 aa Download sequence Send to blast |
MDRAGGPATG LNRTGKSCRL RWVNYLHPGL KHGRMSPKEE HLIIELHARW GNRWSRIARR 60 LPGRTDNEIK NYWRTHMRKK AQERRGDMSP SSSSSSLVYQ SCLLDTVPII SMDGGDIHDD 120 RSCMARVLKS TQSVMDGYTM DQIWKEIEAP GAPSLLGIDE GKDKACSNLP CPLLTSTMSD 180 YSCPEVFWKI DNEETRMLAT QSGYGK* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1gv2_A | 5e-17 | 13 | 81 | 37 | 105 | MYB PROTO-ONCOGENE PROTEIN |
| 1h88_C | 1e-16 | 13 | 81 | 91 | 159 | MYB PROTO-ONCOGENE PROTEIN |
| 1h89_C | 1e-16 | 13 | 81 | 91 | 159 | MYB PROTO-ONCOGENE PROTEIN |
| 1h8a_C | 1e-16 | 13 | 81 | 60 | 128 | MYB TRANSFORMING PROTEIN |
| 1mse_C | 5e-17 | 13 | 81 | 37 | 105 | C-Myb DNA-Binding Domain |
| 1msf_C | 5e-17 | 13 | 81 | 37 | 105 | C-Myb DNA-Binding Domain |
| Search in ModeBase | ||||||
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Os.10174 | 0.0 | leaf| panicle| root| stem | ||||
| Expression -- Microarray ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | ID | |||||
| Expression Atlas | H2KWK6 | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor. {ECO:0000305}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00600 | PBM | Transfer from AT5G59780 | Download |
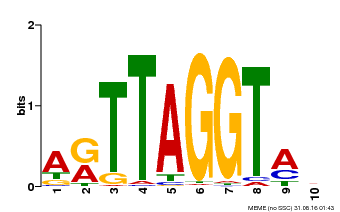
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | LOC_Os11g47460.2 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Phenotype -- Mutation ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | ID | |||||
| RiceGE | Os11g47460 | |||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | DQ075259 | 0.0 | DQ075259.1 Oryza sativa (japonica cultivar-group) MYB transcription factor MYBAS1-2 mRNA, complete cds, alternatively spliced. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015617959.1 | 1e-146 | myb-related protein MYBAS1 isoform X2 | ||||
| Swissprot | Q53NK6 | 1e-147 | MYBA1_ORYSJ; Myb-related protein MYBAS1 | ||||
| TrEMBL | H2KWK6 | 1e-152 | H2KWK6_ORYSJ; Myb family transcription factor, putative, expressed | ||||
| STRING | ORUFI11G25190.1 | 1e-145 | (Oryza rufipogon) | ||||
| STRING | OS11T0700500-01 | 1e-145 | (Oryza sativa) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G59780.3 | 1e-51 | myb domain protein 59 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | LOC_Os11g47460.2 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



