 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | LOC_Os12g01490.1 | ||||||||
| Common Name | LOC4351260, Os12g0105600, OsJ_34927, OSNPB_120105600 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza; Oryza sativa
|
||||||||
| Family | G2-like | ||||||||
| Protein Properties | Length: 315aa MW: 34550.1 Da PI: 7.8679 | ||||||||
| Description | G2-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 97.9 | 7e-31 | 57 | 110 | 2 | 55 |
G2-like 2 prlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRl 55
prlrWtpeLH +Fv+av++LGG+++AtPk +l+lm+v gL++ hvkSHLQ+YR+
LOC_Os12g01490.1 57 PRLRWTPELHLSFVRAVDRLGGQDRATPKLVLQLMNVRGLSIGHVKSHLQMYRS 110
9****************************************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 11.92 | 53 | 113 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 5.6E-29 | 53 | 111 | IPR009057 | Homeodomain-like |
| SuperFamily | SSF46689 | 3.23E-15 | 54 | 111 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 1.2E-22 | 56 | 112 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 1.2E-8 | 58 | 109 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Plant Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PO Term | PO Category | PO Description | ||||
| PO:0020039 | anatomy | leaf lamina | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 315 aa Download sequence Send to blast |
MEKTSAGKEE GSESKTAAAN NDGSSTSSTT EEEESGESQR RTSSSSSVRP YIRSKNPRLR 60 WTPELHLSFV RAVDRLGGQD RATPKLVLQL MNVRGLSIGH VKSHLQMYRS KKIDESGQVI 120 GGGSWRSSDE QQYHHLQMQG GGDGGQAYNL GHLSLPAALH HRHITAGSGT ILQSRVANAW 180 SPWRCHGSYW LRAGHHLLVG SKPYYPPPPA EARANTSSNH PDFVQGSSSS PDDHTMNHQR 240 PVVLKEMIYN EGSNHQGGPL NLDLSLDICP RGEKRKRELI SWRKHEEDHD HTTIAIGGDQ 300 EAESCATGLS LSLF* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6j4k_A | 4e-18 | 58 | 112 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4k_B | 4e-18 | 58 | 112 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_A | 4e-18 | 58 | 112 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_C | 4e-18 | 58 | 112 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_D | 4e-18 | 58 | 112 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_F | 4e-18 | 58 | 112 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_H | 4e-18 | 58 | 112 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_J | 4e-18 | 58 | 112 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| Search in ModeBase | ||||||
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Os.18448 | 0.0 | callus| leaf| panicle| root | ||||
| Expression -- Microarray ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | ID | E-value | ||||
| GEO | 32992506 | 0.0 | ||||
| Expression Atlas | Q2QYV2 | - | ||||
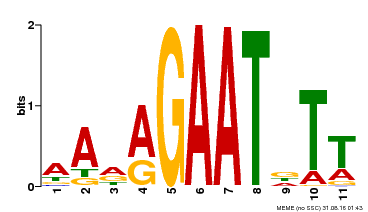
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00297 | DAP | Transfer from AT2G38300 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | LOC_Os12g01490.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Phenotype -- Mutation ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | ID | |||||
| RiceGE | Os12g01490 | |||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK107297 | 0.0 | AK107297.1 Oryza sativa Japonica Group cDNA clone:002-126-C05, full insert sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015620339.1 | 0.0 | probable transcription factor RL9 | ||||
| TrEMBL | Q2QYV2 | 0.0 | Q2QYV2_ORYSJ; Myb-like DNA-binding domain, SHAQKYF class family protein, expressed | ||||
| STRING | OS12T0105600-01 | 0.0 | (Oryza sativa) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP4408 | 36 | 71 | Representative plant | OGRP285 | 15 | 123 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G38300.1 | 2e-38 | G2-like family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | LOC_Os12g01490.1 |
| Entrez Gene | 4351260 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



