 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | LPERR01G07340.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Leersia
|
||||||||
| Family | ERF | ||||||||
| Protein Properties | Length: 357aa MW: 38309.4 Da PI: 4.2767 | ||||||||
| Description | ERF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 62 | 1.3e-19 | 130 | 181 | 1 | 56 |
AP2 1 sgykGVrwdkkrgrWvAeIrd.psengkr.krfslgkfgtaeeAakaaiaarkklege 56
++++GVr+++ +g+++AeIrd + r r++lg+f+taeeAak ++ a+ +l+g+
LPERR01G07340.1 130 PRFRGVRRRP-WGKYAAEIRDpW-----RrVRVWLGTFDTAEEAAKVYDSAAIQLRGA 181
689*******.**********88.....55*************************985 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| CDD | cd00018 | 2.01E-29 | 130 | 187 | No hit | No description |
| Pfam | PF00847 | 4.2E-12 | 130 | 180 | IPR001471 | AP2/ERF domain |
| SuperFamily | SSF54171 | 9.15E-21 | 131 | 189 | IPR016177 | DNA-binding domain |
| SMART | SM00380 | 9.1E-31 | 131 | 193 | IPR001471 | AP2/ERF domain |
| PROSITE profile | PS51032 | 22.958 | 131 | 188 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 1.5E-28 | 131 | 189 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 4.6E-10 | 132 | 143 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 4.6E-10 | 154 | 170 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0042991 | Biological Process | transcription factor import into nucleus | ||||
| GO:0048364 | Biological Process | root development | ||||
| GO:0048825 | Biological Process | cotyledon development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0042802 | Molecular Function | identical protein binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 357 aa Download sequence Send to blast |
MEAEEAFVAV KRTEHVEVTS LIVDSSAAKG NKGKVVAAAA AAGVMPARVR VFCDDVDATD 60 SSSDEEDDEE EVTVRRRVKR YVQEIRLQRA TALPVKAEVL PPVVSTAKAA ATAVVLSGRK 120 RKAEGGGGEP RFRGVRRRPW GKYAAEIRDP WRRVRVWLGT FDTAEEAAKV YDSAAIQLRG 180 ADATTNFNRS GDLLADVPPE VAKRVPQPPE AEKNASPATS YDSGEESHAA AAASPTSVLR 240 SFPPSAVDKK PSPSPPLVVR ESDESSGGFF GCSFSDDGGY AGELPPLYSD FDLLADFPEP 300 PLDFLSNLPD EPFSLAPFSD DDDGDDGDAP SPAAEQQVDD FFQDITDLFQ IDPLPVV |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5wx9_A | 1e-18 | 131 | 187 | 14 | 71 | Ethylene-responsive transcription factor ERF096 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00027 | PBM | Transfer from AT4G23750 | Download |
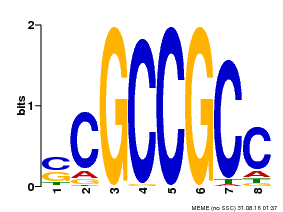
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | LPERR01G07340.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | - | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK101054 | 1e-107 | AK101054.1 Oryza sativa Japonica Group cDNA clone:J033015K21, full insert sequence. | |||
| GenBank | AK106517 | 1e-107 | AK106517.1 Oryza sativa Japonica Group cDNA clone:002-107-B02, full insert sequence. | |||
| GenBank | AP001800 | 1e-107 | AP001800.2 Oryza sativa Japonica Group genomic DNA, chromosome 1, PAC clone:P0443E05. | |||
| GenBank | AP002835 | 1e-107 | AP002835.1 Oryza sativa Japonica Group genomic DNA, chromosome 1, PAC clone:P0417G05. | |||
| GenBank | AP014957 | 1e-107 | AP014957.1 Oryza sativa Japonica Group DNA, chromosome 1, cultivar: Nipponbare, complete sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015629650.1 | 1e-156 | ethylene-responsive transcription factor CRF2 | ||||
| TrEMBL | A0A0D9UYG4 | 0.0 | A0A0D9UYG4_9ORYZ; Uncharacterized protein | ||||
| STRING | LPERR01G07340.1 | 0.0 | (Leersia perrieri) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP1690 | 36 | 97 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G23750.2 | 9e-32 | cytokinin response factor 2 | ||||



