 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | LPERR01G10950.2 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Leersia
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 720aa MW: 76360.3 Da PI: 8.0872 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 49.2 | 9.4e-16 | 498 | 544 | 4 | 55 |
HHHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 4 ahnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
hn ErrRRdriN+++ L+el+P++ K +Ka++L +A+eY+k Lq
LPERR01G10950.2 498 VHNLSERRRRDRINEKMRALQELIPNC-----NKIDKASMLDEAIEYLKTLQ 544
5*************************8.....6******************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.280.10 | 1.8E-19 | 491 | 550 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 3.61E-17 | 491 | 548 | No hit | No description |
| SuperFamily | SSF47459 | 4.45E-20 | 492 | 558 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS50888 | 17.981 | 494 | 543 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 3.2E-13 | 498 | 544 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 6.3E-17 | 500 | 549 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009704 | Biological Process | de-etiolation | ||||
| GO:0009740 | Biological Process | gibberellic acid mediated signaling pathway | ||||
| GO:0010017 | Biological Process | red or far-red light signaling pathway | ||||
| GO:0031539 | Biological Process | positive regulation of anthocyanin metabolic process | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0042802 | Molecular Function | identical protein binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 720 aa Download sequence Send to blast |
MRRLSLLIYL LVHSVSPRSL LGPPTQPRRL SLLFSLIPHA DQQFSAARDF SLPRFTFYFL 60 SLLFSFGFGF HERELTLTSH PAQLRKHGLD CIRLVGVLQL QTRSPSLLDL FSLLGLQAQM 120 SDGNDFAELL WENGQAVVHG RRKHAPPAFP PFGFGGGGGG GSSSNRAQER HPGGGDAFAK 180 AGNFVALGMG AAVHDFTPTF GGAAHDNGDD DAVPWIHYPI IDDDDGAAAP ALAANDYGSD 240 FFSELQAAAA AAAAAAAPPN DLGSLPTNNR NAPIATTCSR EPPSKETHGG GLSVPTTQAS 300 PQLAAGAAKL PRPSGSGSGD GVMNFSLFSR PAVLARATLQ SAQRTQGTDK ASNVTAASNR 360 VESTVVQTAS GARSGPAFAA DQRAWPPQPK EMPFASTAAA APVATAVNLQ HEIGSDRAGR 420 NIGVHKNEGR KAPEATVATS SVCSGNGAGN DELWRQHKRK RQAQAECSAS QEDDIDDEPS 480 ALRRSATRST KRSRTAEVHN LSERRRRDRI NEKMRALQEL IPNCNKIDKA SMLDEAIEYL 540 KTLQLQVQLS LTQMMSMGTG LCIPPMLLPA AMQHLQIPPM AHFPHLGMGL GYGMGVFDMN 600 NTGTLPMPHM PGAHFPCPMI PGASPQGLGI PGTNTLPMFG VPGQAVPSSA PSIPPFPSLA 660 GLHVRPNGVP QVSGAMANMV QDQQQGIGNQ QQQCLNKEAI QGANTNDPQM QIIMQVLINK |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 502 | 507 | ERRRRD |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor that may act as negative regulator of phyB-dependent light signal transduction. {ECO:0000269|PubMed:17485859}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00081 | ChIP-seq | Transfer from AT1G09530 | Download |
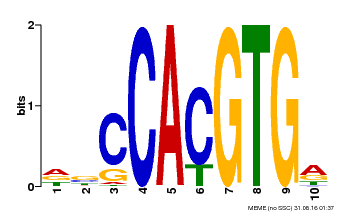
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | LPERR01G10950.2 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Down-regulated by light in dark-grown etiolated seedlings. {ECO:0000269|PubMed:17485859}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK102252 | 0.0 | AK102252.1 Oryza sativa Japonica Group cDNA clone:J033088H11, full insert sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015619027.1 | 0.0 | transcription factor PHYTOCHROME INTERACTING FACTOR-LIKE 15-like isoform X1 | ||||
| Swissprot | Q0JNI9 | 0.0 | PIL15_ORYSJ; Transcription factor PHYTOCHROME INTERACTING FACTOR-LIKE 15 | ||||
| TrEMBL | A0A0D9UZU4 | 0.0 | A0A0D9UZU4_9ORYZ; Uncharacterized protein | ||||
| STRING | LPERR01G10950.2 | 0.0 | (Leersia perrieri) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP3527 | 37 | 75 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G09530.2 | 8e-25 | phytochrome interacting factor 3 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



