 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | LPERR01G34430.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Leersia
|
||||||||
| Family | LBD | ||||||||
| Protein Properties | Length: 255aa MW: 26319.6 Da PI: 8.0505 | ||||||||
| Description | LBD family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | DUF260 | 137 | 6.8e-43 | 32 | 130 | 1 | 99 |
DUF260 1 aCaaCkvlrrkCakdCvlapyfpaeqpkkfanvhklFGasnvlkllkalpeeeredamsslvyeAearardPvyGavgvilklqqqleqlkael 94
+CaaCk+lrrkC++dCv+apyfp ++p+kf vh++FGasnv+kll++l++ +reda++sl+yeA++r+rdPvyG+v++i+ lq++l+ql+++l
LPERR01G34430.1 32 PCAACKFLRRKCQPDCVFAPYFPPDNPQKFVHVHRVFGASNVTKLLNELHPYQREDAVNSLAYEADMRLRDPVYGCVAIISILQRNLRQLQQDL 125
7********************************************************************************************* PP
DUF260 95 allke 99
a++k
LPERR01G34430.1 126 ARAKF 130
99875 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50891 | 26.332 | 31 | 132 | IPR004883 | Lateral organ boundaries, LOB |
| Pfam | PF03195 | 1.2E-42 | 32 | 128 | IPR004883 | Lateral organ boundaries, LOB |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009799 | Biological Process | specification of symmetry | ||||
| GO:0009944 | Biological Process | polarity specification of adaxial/abaxial axis | ||||
| GO:0009954 | Biological Process | proximal/distal pattern formation | ||||
| GO:0048441 | Biological Process | petal development | ||||
| GO:0005654 | Cellular Component | nucleoplasm | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 255 aa Download sequence Send to blast |
MASSSASSVP APSGSVITIA SAAAATCGTG SPCAACKFLR RKCQPDCVFA PYFPPDNPQK 60 FVHVHRVFGA SNVTKLLNEL HPYQREDAVN SLAYEADMRL RDPVYGCVAI ISILQRNLRQ 120 LQQDLARAKF ELSKYQVAAA AANNGMAEFI GNAVPNGQSF INVGHTAALA SIGGSAACFG 180 QEQFSTVQML SAARSYEGEP IARLGGNGGY EFGYSTAAMA GGGAGHMSGL GTLGGGPFLK 240 SGIAGSDETR SGAGQ |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5ly0_A | 1e-54 | 22 | 135 | 1 | 114 | LOB family transfactor Ramosa2.1 |
| 5ly0_B | 1e-54 | 22 | 135 | 1 | 114 | LOB family transfactor Ramosa2.1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Negative regulator of cell proliferation in the adaxial side of leaves. Regulates the formation of a symmetric lamina and the establishment of venation (By similarity). {ECO:0000250}. | |||||
| UniProt | Negative regulator of cell proliferation in the adaxial side of leaves. Regulates the formation of a symmetric lamina and the establishment of venation (By similarity). {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00211 | DAP | Transfer from AT1G65620 | Download |
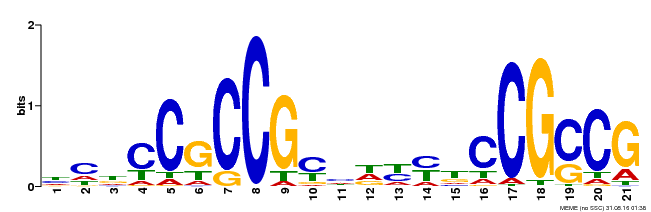
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | LPERR01G34430.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AB200237 | 1e-150 | AB200237.1 Oryza sativa Japonica Group CRLL3 mRNA for LOB domain protein, complete cds. | |||
| GenBank | AK071407 | 1e-150 | AK071407.1 Oryza sativa Japonica Group cDNA clone:J023089G14, full insert sequence. | |||
| GenBank | AK119575 | 1e-150 | AK119575.1 Oryza sativa Japonica Group cDNA clone:002-117-B04, full insert sequence. | |||
| GenBank | EF540766 | 1e-150 | EF540766.1 Oryza sativa Japonica Group LOB domain protein 6 mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015634701.1 | 1e-141 | LOB domain-containing protein 6 | ||||
| Swissprot | A2WXT0 | 1e-142 | LBD6_ORYSI; LOB domain-containing protein 6 | ||||
| Swissprot | Q8LQH4 | 1e-142 | LBD6_ORYSJ; LOB domain-containing protein 6 | ||||
| TrEMBL | A0A0D9V8S9 | 0.0 | A0A0D9V8S9_9ORYZ; Uncharacterized protein | ||||
| STRING | LPERR01G34430.1 | 0.0 | (Leersia perrieri) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP2267 | 38 | 95 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G65620.4 | 2e-50 | LBD family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



