 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | LPERR01G36170.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Leersia
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 473aa MW: 49944.8 Da PI: 6.6456 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 29.3 | 1.6e-09 | 279 | 326 | 4 | 55 |
HHHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 4 ahnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
+h+ +Er RR++i +++ L++l+P + +k + Ka +L + ++Y++sLq
LPERR01G36170.1 279 SHSLAERVRREKISQRMKLLQDLVPGC----NKVVGKAVMLDEIINYVQSLQ 326
8*************************9....677*****************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| CDD | cd00083 | 3.16E-10 | 273 | 330 | No hit | No description |
| SuperFamily | SSF47459 | 1.31E-16 | 273 | 344 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS50888 | 15.256 | 275 | 325 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene3D | G3DSA:4.10.280.10 | 1.1E-16 | 277 | 343 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 1.4E-6 | 279 | 326 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 1.3E-10 | 281 | 331 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 473 aa Download sequence Send to blast |
MNCGPPDQLP PATAPSCFLN LNWDQSMDPA LSSMVSSPAS NSTAAPPDGL ALHGISPQQP 60 HYGGGTPLSS PPKLNLSMMG QFHPYPPPPP GAGGLPILEN PMPMGHLDQF LADPGFAERA 120 ARLSGFDARG GGGGYGGAAV QPQFGLPDAS PAGGSKEMEL GNTRDESSVS GPAPGGGEIP 180 LKGASDGNAR KRKASGKGKS KDSPMSTSAS KEDSSGKRCK STEERENNAA GEENSGKGKV 240 AQSNSENGKK GGKESSSKPP EPPKDYIHVR ARRGEATDSH SLAERVRREK ISQRMKLLQD 300 LVPGCNKVVG KAVMLDEIIN YVQSLQRQVE FLSMKLATVN PQLDFNNLPN LLAKDMHQSC 360 SPLQSSHFPL ETSGAPLPYI NQPQQGNPLG CGLTNGMDNQ GSMHPLDQAF CRQMGSHHPF 420 LNGVSDAASQ VGAFWQDDLQ SVVQMDMGQS QEIATSSNSY NGSLQTVHMK MEL |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00660 | PBM | Transfer from Pp3c6_9520 | Download |
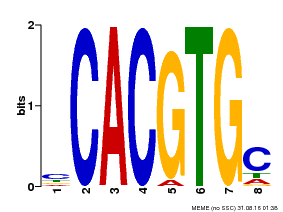
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | LPERR01G36170.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | - | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK101063 | 0.0 | AK101063.1 Oryza sativa Japonica Group cDNA clone:J033022O19, full insert sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015621927.1 | 0.0 | transcription factor bHLH77 isoform X2 | ||||
| TrEMBL | A0A0D9V9H2 | 0.0 | A0A0D9V9H2_9ORYZ; Uncharacterized protein | ||||
| STRING | LPERR01G36170.2 | 0.0 | (Leersia perrieri) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G07340.1 | 4e-62 | bHLH family protein | ||||



