 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | LPERR03G04660.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Leersia
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 349aa MW: 37701.8 Da PI: 6.7233 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 56.6 | 4.3e-18 | 127 | 180 | 3 | 56 |
--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHHH CS
Homeobox 3 kRttftkeqleeLeelFeknrypsaeereeLAkklgLterqVkvWFqNrRakek 56
k+++++ eq+++Le+ Fe +++ e++ +LA+ lgL+ rqV +WFqNrRa++k
LPERR03G04660.1 127 KKRRLNVEQVRTLEKNFELGNKLEPERKMQLARALGLQPRQVAIWFQNRRARWK 180
4568999**********************************************9 PP
| |||||||
| 2 | HD-ZIP_I/II | 129.4 | 1.4e-41 | 126 | 217 | 1 | 92 |
HD-ZIP_I/II 1 ekkrrlskeqvklLEesFeeeekLeperKvelareLglqprqvavWFqnrRARtktkqlEkdyeaLkraydalkeenerLekeveeLreelk 92
ekkrrl+ eqv++LE++Fe +kLeperK++lar+Lglqprqva+WFqnrRAR+ktkqlEkdy+aLkr++da+k++n++L +++++L++e+
LPERR03G04660.1 126 EKKRRLNVEQVRTLEKNFELGNKLEPERKMQLARALGLQPRQVAIWFQNRRARWKTKQLEKDYDALKRQLDAVKADNDALLSHNKKLQAEIV 217
69**************************************************************************************9986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.60 | 2.3E-19 | 99 | 183 | IPR009057 | Homeodomain-like |
| SuperFamily | SSF46689 | 9.84E-20 | 116 | 184 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50071 | 17.022 | 122 | 182 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 1.2E-17 | 125 | 186 | IPR001356 | Homeobox domain |
| CDD | cd00086 | 6.59E-17 | 127 | 183 | No hit | No description |
| Pfam | PF00046 | 2.0E-15 | 127 | 180 | IPR001356 | Homeobox domain |
| PRINTS | PR00031 | 1.2E-5 | 153 | 162 | IPR000047 | Helix-turn-helix motif |
| PROSITE pattern | PS00027 | 0 | 157 | 180 | IPR017970 | Homeobox, conserved site |
| PRINTS | PR00031 | 1.2E-5 | 162 | 178 | IPR000047 | Helix-turn-helix motif |
| Pfam | PF02183 | 3.6E-14 | 182 | 221 | IPR003106 | Leucine zipper, homeobox-associated |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009744 | Biological Process | response to sucrose | ||||
| GO:0048826 | Biological Process | cotyledon morphogenesis | ||||
| GO:0080022 | Biological Process | primary root development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 349 aa Download sequence Send to blast |
MASNGMASSP SSFFPPNFLL HMAQQQAAAA PTPTHHDLEE HHHHEQHHHH HHHLGLPPPA 60 PPPHPNHNPF LPSSQCQSLQ EFRGMAPMLS VGTKRPMYPE SAGEEMNGGG EDELSDDGSQ 120 AGGGGEKKRR LNVEQVRTLE KNFELGNKLE PERKMQLARA LGLQPRQVAI WFQNRRARWK 180 TKQLEKDYDA LKRQLDAVKA DNDALLSHNK KLQAEIVALK GSREAASELI NLNKETEASC 240 SNRSENSSEI NLDISRTPPA DGGGGGGMIP FYPSARPGAG AGAGVDIDHL LHTSSAAAKM 300 EHHGGGGGGG GGGNNVQASV DTASFGNLLC GAVDEPPPFW PWPDHQHFH |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 174 | 182 | RRARWKTKQ |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00225 | DAP | Transfer from AT1G69780 | Download |
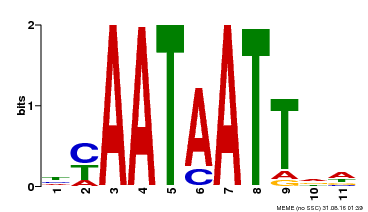
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | LPERR03G04660.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | BT088234 | 1e-147 | BT088234.1 Zea mays full-length cDNA clone ZM_BFc0086K06 mRNA, complete cds. | |||
| GenBank | JX469955 | 1e-147 | JX469955.1 Zea mays subsp. mays clone UT3217 HB-type transcription factor mRNA, partial cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_002465782.1 | 1e-153 | homeobox-leucine zipper protein HOX21 | ||||
| Swissprot | Q8S7W9 | 1e-150 | HOX21_ORYSJ; Homeobox-leucine zipper protein HOX21 | ||||
| TrEMBL | A0A0D9VQ14 | 0.0 | A0A0D9VQ14_9ORYZ; Uncharacterized protein | ||||
| STRING | LPERR03G04660.1 | 0.0 | (Leersia perrieri) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP2289 | 38 | 94 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G69780.1 | 3e-72 | HD-ZIP family protein | ||||



