 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | LPERR03G05920.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Leersia
|
||||||||
| Family | CAMTA | ||||||||
| Protein Properties | Length: 988aa MW: 110744 Da PI: 5.783 | ||||||||
| Description | CAMTA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | CG-1 | 177.5 | 1.7e-55 | 19 | 136 | 2 | 118 |
CG-1 2 lke.kkrwlkneeiaaiLenfekheltlelktrpksgsliLynrkkvryfrkDGyswkkkkdgktvrEdhekLKvggvevlycyYahseenptf 94
lke ++rwl+++ei++iL+n+++++++ e+++rp+sgsl+L++rk++ryfrkDG++w+kk+dgktv+E+he+LK g+++vl+cyYah+een +f
LPERR03G05920.1 19 LKEaQQRWLRPTEICEILKNYRNFRIAPEPPNRPPSGSLFLFDRKVLRYFRKDGHNWRKKRDGKTVKEAHERLKSGSIDVLHCYYAHGEENINF 112
45559***************************************************************************************** PP
CG-1 95 qrrcywlLeeelekivlvhylevk 118
qrr+yw+Lee++ +ivlvhylevk
LPERR03G05920.1 113 QRRSYWMLEEDYMHIVLVHYLEVK 136
*********************985 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51437 | 82.118 | 15 | 141 | IPR005559 | CG-1 DNA-binding domain |
| SMART | SM01076 | 6.6E-76 | 18 | 136 | IPR005559 | CG-1 DNA-binding domain |
| Pfam | PF03859 | 1.1E-48 | 21 | 135 | IPR005559 | CG-1 DNA-binding domain |
| Gene3D | G3DSA:2.60.40.10 | 7.9E-6 | 401 | 486 | IPR013783 | Immunoglobulin-like fold |
| Pfam | PF01833 | 1.3E-6 | 402 | 479 | IPR002909 | IPT domain |
| SuperFamily | SSF81296 | 9.97E-16 | 402 | 487 | IPR014756 | Immunoglobulin E-set |
| SuperFamily | SSF48403 | 4.35E-17 | 590 | 694 | IPR020683 | Ankyrin repeat-containing domain |
| CDD | cd00204 | 5.82E-14 | 593 | 691 | No hit | No description |
| Gene3D | G3DSA:1.25.40.20 | 4.5E-18 | 593 | 701 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50088 | 8.79 | 599 | 631 | IPR002110 | Ankyrin repeat |
| PROSITE profile | PS50297 | 18.165 | 599 | 703 | IPR020683 | Ankyrin repeat-containing domain |
| Pfam | PF12796 | 4.0E-6 | 604 | 694 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50088 | 8.843 | 632 | 664 | IPR002110 | Ankyrin repeat |
| SMART | SM00248 | 0.013 | 632 | 661 | IPR002110 | Ankyrin repeat |
| SMART | SM00248 | 1000 | 671 | 700 | IPR002110 | Ankyrin repeat |
| SuperFamily | SSF52540 | 4.51E-8 | 803 | 856 | IPR027417 | P-loop containing nucleoside triphosphate hydrolase |
| SMART | SM00015 | 0.11 | 805 | 827 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 8.261 | 807 | 835 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 0.0032 | 807 | 826 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 0.0022 | 828 | 850 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 9.578 | 829 | 853 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 1.9E-4 | 831 | 850 | IPR000048 | IQ motif, EF-hand binding site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009409 | Biological Process | response to cold | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0071275 | Biological Process | cellular response to aluminum ion | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 988 aa Download sequence Send to blast |
MAEGRRYAIA PQLDIEQILK EAQQRWLRPT EICEILKNYR NFRIAPEPPN RPPSGSLFLF 60 DRKVLRYFRK DGHNWRKKRD GKTVKEAHER LKSGSIDVLH CYYAHGEENI NFQRRSYWML 120 EEDYMHIVLV HYLEVKAGKL SSRGRGNDDI LQASCVDSPL SQLPSQTTEG ESSVSGQASE 180 YDETESGSYQ GLQATTPNTG FYSHGQEKLY VPLNETDFGT AFNGPNSQFD LSLWIETMKP 240 YKGTNQIPLY QALVPSEQSP FTEDQGIESF TFDEVYNNGL SIKDVGGDGT GGETPWQIPN 300 ASGSFATVDS FQQNEKTLEE AINPLLKTHS SSLSDILKDS FKKNDSFTRW MSKELAEVDD 360 SQITSSSGGY WNSEETDDII EASSSDQFTL GPVLAQDQLF SIVDFSPTWT YAGSKTRVFV 420 KGKILNSDEV KRFKWSCMFG EDEVPAEILA DETLMCYSPS HKPGRVPFYV TCSNRLACSE 480 VREFEFRTQY MDAPSPHGST NKTYLQMRLD KLLSLGQNEI QATLSNSTKE IIDLSKKINS 540 LMMNNDDWSE LLKLADDNEP ATDDKQDQFL QNRIKEKLYI WLLHKVGDGG KGPSVLDEEG 600 QGVLHLAAAL GYDWAIRPTI TAGVNINFRD AHGWTALHWA AFCGRERTVV ALIALGAAPG 660 AVTDPSPSFP TGNTPADLAS ANGHKGISGF LAESSLTSHL QTLNLKEAMR SSAAEISGLP 720 GIVNVADTST SPLAGESLQT GSMGDSLGAV RNAAQAAARI FQVFRMQSFQ RKQAVQYEDD 780 NGAISDERAM SLLSAKPSKP AQLDPVHAAA TRIQNKFRGW KGRKEFLLIR QRIVKIQAHV 840 RGHQVRKHYR KIIWSVGIVE KVILRWRRRG AGLRGFRPTE SAVTESTSSS SGDMTQNRPA 900 ENDYDFLQKG RKQTEERLQK ALARVKSMVQ YPDARDQYQR ILTAVTKMQE SQAMQEKMLE 960 ESTEMDEGLL MSEFKELWDD DMPTPGYF |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator that binds to the DNA consensus sequence 5'-[ACG]CGCG[GTC]-3' (By similarity). Regulates transcriptional activity in response to calcium signals (Probable). Binds calmodulin in a calcium-dependent manner (By similarity). Involved in freezing tolerance in association with CAMTA1 and CAMTA3. Contributes together with CAMTA1 and CAMTA3 to the positive regulation of the cold-induced expression of DREB1A/CBF3, DREB1B/CBF1 and DREB1C/CBF2 (PubMed:23581962). Involved together with CAMTA3 and CAMTA4 in the positive regulation of a general stress response (PubMed:25039701). Involved in tolerance to aluminum. Binds to the promoter of ALMT1 transporter and contributes to the positive regulation of aluminum-induced expression of ALMT1 (PubMed:25627216). {ECO:0000250|UniProtKB:Q8GSA7, ECO:0000269|PubMed:23581962, ECO:0000269|PubMed:25039701, ECO:0000269|PubMed:25627216, ECO:0000305|PubMed:11925432}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00043 | PBM | Transfer from AT5G64220 | Download |
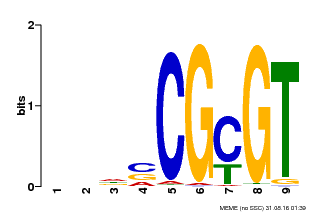
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | LPERR03G05920.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By salt, wounding, abscisic acid, H(2)O(2) and salicylic acid (PubMed:12218065). Induced by aluminum (PubMed:25627216). {ECO:0000269|PubMed:12218065, ECO:0000269|PubMed:25627216}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK066651 | 0.0 | AK066651.1 Oryza sativa Japonica Group cDNA clone:J013074G10, full insert sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015631248.1 | 0.0 | calmodulin-binding transcription activator 1 isoform X1 | ||||
| Swissprot | Q6NPP4 | 0.0 | CMTA2_ARATH; Calmodulin-binding transcription activator 2 | ||||
| TrEMBL | A0A0D9VQK3 | 0.0 | A0A0D9VQK3_9ORYZ; Uncharacterized protein | ||||
| STRING | LPERR03G05920.1 | 0.0 | (Leersia perrieri) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP608 | 38 | 140 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G22300.2 | 0.0 | signal responsive 1 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



