 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | LPERR03G34030.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Leersia
|
||||||||
| Family | SBP | ||||||||
| Protein Properties | Length: 959aa MW: 104723 Da PI: 5.2776 | ||||||||
| Description | SBP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SBP | 129.1 | 1.7e-40 | 141 | 218 | 1 | 78 |
--SSTT-----TT--HHHHHTT--HHHHT-S-EEETTEEEEE-TTTSSEEETTT--SS--S-STTTT-------S--- CS
SBP 1 lCqvegCeadlseakeyhrrhkvCevhskapvvlvsgleqrfCqqCsrfhelsefDeekrsCrrrLakhnerrrkkqa 78
+Cqv+gC+adls +k+yhrrhkvCe+h+ka++++v ++ qrfCqqCsrfh l+efDe+krsCrrrLa+hn+rrrk+++
LPERR03G34030.1 141 TCQVDGCTADLSAVKDYHRRHKVCEMHAKATTCVVGKTVQRFCQQCSRFHLLQEFDEGKRSCRRRLAGHNRRRRKTRP 218
6**************************************************************************875 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.1100.10 | 6.4E-33 | 134 | 203 | IPR004333 | Transcription factor, SBP-box |
| PROSITE profile | PS51141 | 32.231 | 139 | 216 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF103612 | 1.09E-37 | 140 | 221 | IPR004333 | Transcription factor, SBP-box |
| Pfam | PF03110 | 3.8E-29 | 142 | 215 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF48403 | 7.9E-9 | 704 | 851 | IPR020683 | Ankyrin repeat-containing domain |
| CDD | cd00204 | 2.44E-9 | 716 | 846 | No hit | No description |
| Gene3D | G3DSA:1.25.40.20 | 1.4E-8 | 742 | 842 | IPR020683 | Ankyrin repeat-containing domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0016021 | Cellular Component | integral component of membrane | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 959 aa Download sequence Send to blast |
METARVGAQK RLFGWDLNDW RWDSERFVAT PVPTTAAAAA SGLALNSSPS SSEEAEASVP 60 RNVNVRGDFD KRKRVVVIDD DDDDDVEEYE EPVGNGGGSL SLRIGGDAVA HGAGVGGGAG 120 DEEDRNGKKI KVQGGSSSGP TCQVDGCTAD LSAVKDYHRR HKVCEMHAKA TTCVVGKTVQ 180 RFCQQCSRFH LLQEFDEGKR SCRRRLAGHN RRRRKTRPEV AVTGSALAED KVNSYLLLGL 240 LGVCANLNAD SAENLRGQEL LSNLWKNLGA VAKSLDPKEL CKLLEACQSM QDGSNAGTSE 300 TANTLVNTAV AEAAGPSNSK MPFVSGDQCG LASTSVVPVQ SKSPTVATPE PPACKLKDFD 360 LNDTCGDMEG FEDGYEGSPT PAFKTTDSPN CPSWMHQDST QSPPQTSGNS DSTSAQSLSS 420 SNGDAQCRTD KIVFKLFEKV PSDLPPVLRS QILGWLSSSP TDIESYIRPG CIILTVYLRL 480 VESAWKELYD NMSSYLDKLL NSSTGNFWAS GLVFVMVRHQ IAFMHNGQVM LDRPLAISSH 540 HYCKILCVRP IAAPFSTKVN FRVEGFNLVS ASSRLICSFE GCCLFQEDTD NIVDDAEHDD 600 IEYLNFCCPF PGSRGRGFVE VEDGGFSNGF FPFIIAEQEI CSEVCELESV FEPSSHEQAD 660 DDNARNQALE FLNELGWLLH RANRISKQDK FPLASFNIWR FRNLGIFAME RDWCAVTKLL 720 LDFLFIGLVD MGSQSPEEVV LSENLLHAAV RRKSAQMVRF LLGYKPNESL KGTAKTFLFR 780 PDAQGPSKLT PLHIAAATND AEDVLDALTD DPGLVWINAW RSARDDAGFT PEDYACQRGN 840 DAYLNMVEKK INKHLGKGHV VLGVPSSMHP VISDGVKPGE ASLEIGMNVP PQASQCNACS 900 RQALMHPNST ARTFLYRPAM LTVMGIAVVC VCVGLLLHTC PKVYAAPTFR WELLERGAM |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ul4_A | 2e-31 | 135 | 215 | 4 | 84 | squamosa promoter binding protein-like 4 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Trans-acting factor that binds specifically to the consensus nucleotide sequence 5'-TNCGTACAA-3'. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00633 | PBM | Transfer from PK06791.1 | Download |
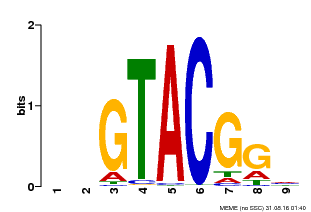
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | LPERR03G34030.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK069847 | 0.0 | AK069847.1 Oryza sativa Japonica Group cDNA clone:J023034C10, full insert sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015631511.1 | 0.0 | squamosa promoter-binding-like protein 6 isoform X1 | ||||
| Swissprot | Q75LH6 | 0.0 | SPL6_ORYSJ; Squamosa promoter-binding-like protein 6 | ||||
| TrEMBL | A0A0D9W117 | 0.0 | A0A0D9W117_9ORYZ; Uncharacterized protein | ||||
| STRING | LPERR03G34030.1 | 0.0 | (Leersia perrieri) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP1998 | 38 | 92 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G47070.1 | 0.0 | squamosa promoter binding protein-like 1 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



