 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | LPERR05G10570.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Leersia
|
||||||||
| Family | ERF | ||||||||
| Protein Properties | Length: 176aa MW: 18703.8 Da PI: 6.5421 | ||||||||
| Description | ERF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 60.3 | 4.4e-19 | 46 | 94 | 3 | 55 |
AP2 3 ykGVrwdkkrgrWvAeIrdpsengkrkrfslgkfgtaeeAakaaiaarkkleg 55
y+G+r+++ +grW+AeIrdp++ +r++lg++ tae+Aa+a++ a++ ++g
LPERR05G10570.1 46 YRGIRRRP-WGRWAAEIRDPRK---GARVWLGTYATAEDAARAYDVAARDIRG 94
9*******.**********954...4***********************9998 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00380 | 1.5E-33 | 45 | 108 | IPR001471 | AP2/ERF domain |
| SuperFamily | SSF54171 | 1.11E-22 | 45 | 104 | IPR016177 | DNA-binding domain |
| Pfam | PF00847 | 4.6E-13 | 45 | 94 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 4.3E-32 | 45 | 102 | IPR001471 | AP2/ERF domain |
| PROSITE profile | PS51032 | 24.724 | 45 | 102 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 4.7E-11 | 46 | 57 | IPR001471 | AP2/ERF domain |
| CDD | cd00018 | 8.22E-28 | 46 | 104 | No hit | No description |
| PRINTS | PR00367 | 4.7E-11 | 68 | 84 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0008219 | Biological Process | cell death | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0009735 | Biological Process | response to cytokinin | ||||
| GO:0009753 | Biological Process | response to jasmonic acid | ||||
| GO:0010286 | Biological Process | heat acclimation | ||||
| GO:0034059 | Biological Process | response to anoxia | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0051707 | Biological Process | response to other organism | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005737 | Cellular Component | cytoplasm | ||||
| GO:0005886 | Cellular Component | plasma membrane | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 176 aa Download sequence Send to blast |
MCGGAIIADF VPSPAARHPT DDDTNFSGEE ESSLAKTAPG GRKTAYRGIR RRPWGRWAAE 60 IRDPRKGARV WLGTYATAED AARAYDVAAR DIRGAKAKLN FPPSIAIAAA HPPPKKRRRS 120 SAAAAAAAES SASSPPAAER QLRECMSGLE AFLGLEEEEG GDGAGEQWDA VDLMLE |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 2gcc_A | 4e-21 | 46 | 105 | 6 | 66 | ATERF1 |
| 3gcc_A | 4e-21 | 46 | 105 | 6 | 66 | ATERF1 |
| 5wx9_A | 4e-21 | 46 | 110 | 15 | 80 | Ethylene-responsive transcription factor ERF096 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probably acts as a transcriptional activator. Binds to the GCC-box pathogenesis-related promoter element. May be involved in the regulation of gene expression by stress factors and by components of stress signal transduction pathways (By similarity). {ECO:0000250}. | |||||
| UniProt | Probably acts as a transcriptional activator. Binds to the GCC-box pathogenesis-related promoter element. May be involved in the regulation of gene expression by stress factors and by components of stress signal transduction pathways (By similarity). {ECO:0000250, ECO:0000269|PubMed:9159183}. | |||||
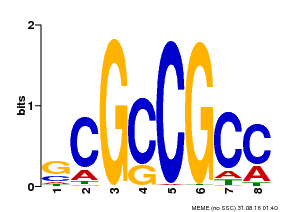
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00109 | PBM | Transfer from AT3G16770 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | LPERR05G10570.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By 1-aminocyclopropane-1-carboxylic acid (ACC, ethylene precursor), methyl jasmonate (MeJA), and Botrytis cinerea. Also induced by cadmium. {ECO:0000269|PubMed:18836139, ECO:0000269|PubMed:9159183}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | - | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AC134348 | 1e-71 | AC134348.2 Oryza sativa Japonica Group cultivar Nipponbare chromosome 5 clone P0530H10, complete sequence. | |||
| GenBank | AC135929 | 1e-71 | AC135929.2 Oryza sativa Japonica Group cultivar Nipponbare chromosome 5 clone P0692D12, complete sequence. | |||
| GenBank | AK100184 | 1e-71 | AK100184.1 Oryza sativa Japonica Group cDNA clone:J023031H12, full insert sequence. | |||
| GenBank | AK111676 | 1e-71 | AK111676.1 Oryza sativa Japonica Group cDNA clone:J013155B14, full insert sequence. | |||
| GenBank | AP014961 | 1e-71 | AP014961.1 Oryza sativa Japonica Group DNA, chromosome 5, cultivar: Nipponbare, complete sequence. | |||
| GenBank | CP012613 | 1e-71 | CP012613.1 Oryza sativa Indica Group cultivar RP Bio-226 chromosome 5 sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006654302.1 | 2e-64 | PREDICTED: ethylene-responsive transcription factor ERF071-like | ||||
| Swissprot | O22259 | 6e-34 | ERF71_ARATH; Ethylene-responsive transcription factor ERF071 | ||||
| Swissprot | P42736 | 4e-33 | RAP23_ARATH; Ethylene-responsive transcription factor RAP2-3 | ||||
| TrEMBL | A0A0D9WFL8 | 1e-121 | A0A0D9WFL8_9ORYZ; Uncharacterized protein | ||||
| STRING | LPERR05G10570.1 | 1e-122 | (Leersia perrieri) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP103 | 37 | 465 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G47520.1 | 4e-23 | ERF family protein | ||||



