 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | LPERR05G12780.2 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Leersia
|
||||||||
| Family | SBP | ||||||||
| Protein Properties | Length: 846aa MW: 93105.6 Da PI: 5.9117 | ||||||||
| Description | SBP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SBP | 111.5 | 5e-35 | 170 | 245 | 1 | 76 |
--SSTT-----TT--HHHHHTT--HHHHT-S-EEETTEEEEE-TTTSSEEETTT--SS--S-STTTT-------S- CS
SBP 1 lCqvegCeadlseakeyhrrhkvCevhskapvvlvsgleqrfCqqCsrfhelsefDeekrsCrrrLakhnerrrkk 76
+Cqv+gCead+ e+k yhrrh+vC +++a++v+++g+++r+CqqC++fh l +fDe+krsCrr+L++hn+rrr+k
LPERR05G12780.2 170 RCQVPGCEADIRELKGYHRRHRVCLRCAHAAAVMLDGVQKRYCQQCGKFHILLDFDEDKRSCRRKLERHNKRRRRK 245
6************************************************************************997 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.1100.10 | 2.6E-27 | 165 | 232 | IPR004333 | Transcription factor, SBP-box |
| PROSITE profile | PS51141 | 29.581 | 168 | 245 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF103612 | 1.57E-33 | 169 | 248 | IPR004333 | Transcription factor, SBP-box |
| Pfam | PF03110 | 6.9E-27 | 171 | 245 | IPR004333 | Transcription factor, SBP-box |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0055070 | Biological Process | copper ion homeostasis | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0016021 | Cellular Component | integral component of membrane | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 846 aa Download sequence Send to blast |
MDAPSAGTSA GGGAAGDAGE PVWDWGNLLD FAIQDDDESL VLPWDGSIGI EADPTTEATL 60 TLPSAPPSSQ PVEAAAEAAP PPLTMQPEGS KRRVRKRDPR LVCPNYLAGR VPCACPEVDE 120 MAAALEVEDV ATDLLAGARK KSKSVGRGGG DAAAGGSGGG ASRGTPAEMR CQVPGCEADI 180 RELKGYHRRH RVCLRCAHAA AVMLDGVQKR YCQQCGKFHI LLDFDEDKRS CRRKLERHNK 240 RRRRKPDSKG ILEKDVDDQL DLSADGSGDV GSGDGELREE NMDGTASETL ETVLSNKVLD 300 KETPVGSEDV LSSPTCAQPS FQIDQSKSLV TFAASVEACL GAKQENNKLS NSPVHDTKST 360 YSSSCPTGRL SFKLYDWNPA EFPRRLRHQI FEWLSSMPVE LEGYIRPGCT ILTVFVAMPQ 420 HMWDKLSEDT ATLVKSLINA PNSLLLGKGA FFVHVNNMIF QVLKDGATLT SARLDVQSPR 480 IHYVYPSWFE TGKPVELILC GSSLDQPKFR SLLSFNGLYL KHDCCRIMPH ETFDYVESRE 540 SILDSQHEIF RINITQSKLD THGPAFVEVE NMFGLSNFVP ILVGSKDLCS ELEQIHDALC 600 GSSDKSSDPC DLRGFRQTGM SGFLIDIGWL IRKPSRDEFQ NLLSLPNIQR WICMMKFLIR 660 NNYINVLKII VNSLENIIGS EVLSNLEKGR LENHVTEFLG YVSQARNIVD SRPKYDKQTQ 720 IDTRWEVDNA PNQPNLGSSV PLVEESTSGE HDLHPTNEAS GEEENMPLVT KALPHRQCCH 780 PEITTRWLNP ASVGVFPGGA IRMRLATTVV IAAVLCFTAC IVLFHPNRVG VIAAPVKRYL 840 SRNYSL |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ul5_A | 1e-37 | 170 | 250 | 5 | 85 | squamosa promoter binding protein-like 7 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 227 | 245 | KRSCRRKLERHNKRRRRKP |
| 2 | 231 | 243 | RRKLERHNKRRRR |
| 3 | 238 | 245 | NKRRRRKP |
| 4 | 239 | 244 | KRRRRK |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Trans-acting factor that binds specifically to the consensus nucleotide sequence 5'-TNCGTACAA-3'. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00604 | PBM | Transfer from AT5G18830 | Download |
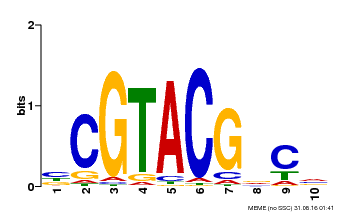
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | LPERR05G12780.2 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK100057 | 0.0 | AK100057.1 Oryza sativa Japonica Group cDNA clone:J013158B16, full insert sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015640052.1 | 0.0 | squamosa promoter-binding-like protein 9 | ||||
| Swissprot | Q6I576 | 0.0 | SPL9_ORYSJ; Squamosa promoter-binding-like protein 9 | ||||
| TrEMBL | A0A0D9WGE5 | 0.0 | A0A0D9WGE5_9ORYZ; Uncharacterized protein | ||||
| STRING | LPERR05G12780.2 | 0.0 | (Leersia perrieri) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP6922 | 33 | 44 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G18830.2 | 1e-149 | squamosa promoter binding protein-like 7 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



