 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | LPERR07G12350.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Leersia
|
||||||||
| Family | CAMTA | ||||||||
| Protein Properties | Length: 859aa MW: 95744.9 Da PI: 8.4054 | ||||||||
| Description | CAMTA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | CG-1 | 99.7 | 2.2e-31 | 32 | 106 | 4 | 77 |
CG-1 4 e.kkrwlkneeiaaiLenfekheltlelktrpksgsliLynrkkvryfrkDGyswkkkkdgktvrEdhekLKvgg 77
e rw +++ei+aiL+n++++++++++ ++p sg+++Ly+rk+vr+frkDG+swkkkkdg+tv+E+hekLK ++
LPERR07G12350.1 32 EaAVRWFRPNEIYAILANHARFKIHAQPVDKPVSGTVVLYDRKVVRNFRKDGHSWKKKKDGRTVQEAHEKLKENA 106
4488********************************************************************765 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51437 | 41.187 | 26 | 154 | IPR005559 | CG-1 DNA-binding domain |
| SMART | SM01076 | 9.4E-26 | 29 | 213 | IPR005559 | CG-1 DNA-binding domain |
| Pfam | PF03859 | 4.4E-25 | 33 | 105 | IPR005559 | CG-1 DNA-binding domain |
| SuperFamily | SSF81296 | 3.97E-11 | 323 | 409 | IPR014756 | Immunoglobulin E-set |
| SuperFamily | SSF48403 | 2.18E-13 | 499 | 609 | IPR020683 | Ankyrin repeat-containing domain |
| CDD | cd00204 | 1.31E-11 | 507 | 603 | No hit | No description |
| Gene3D | G3DSA:1.25.40.20 | 7.5E-14 | 523 | 609 | IPR020683 | Ankyrin repeat-containing domain |
| Pfam | PF12796 | 9.5E-7 | 532 | 607 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50297 | 14.902 | 535 | 615 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50088 | 11.781 | 544 | 576 | IPR002110 | Ankyrin repeat |
| SMART | SM00248 | 6.4E-6 | 544 | 573 | IPR002110 | Ankyrin repeat |
| SMART | SM00248 | 580 | 583 | 612 | IPR002110 | Ankyrin repeat |
| SuperFamily | SSF52540 | 5.31E-9 | 660 | 761 | IPR027417 | P-loop containing nucleoside triphosphate hydrolase |
| SMART | SM00015 | 0.83 | 710 | 732 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 7.657 | 711 | 737 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 0.0026 | 713 | 731 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 0.0095 | 733 | 755 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 10.493 | 734 | 758 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 0.0047 | 735 | 755 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 2.4 | 816 | 838 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 9.084 | 818 | 846 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 0.04 | 819 | 838 | IPR000048 | IQ motif, EF-hand binding site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 859 aa Download sequence Send to blast |
MAGAGGWDAL IGSEIHGFLT YADLNYEKLV AEAAVRWFRP NEIYAILANH ARFKIHAQPV 60 DKPVSGTVVL YDRKVVRNFR KDGHSWKKKK DGRTVQEAHE KLKENAMAAP NSEPEAADLP 120 TVNLVHTSPL TSADSTSAHT EQSYSTAVPE EINSHGGISA SSETSNHDSS LEEFWANLLE 180 SSIKNDPKSG SFSSSQQINN GPKNSENIIF NTSMSSNAIP ALYMASETYP TNHGLNQVTA 240 DHFKALKNQG DQTHSLFTSN VDFQSDQFIS SLVKSSMDGK VSDPNDVPAR QNSLGLWKYL 300 DDDNPGLEDN PGSVTQTNKT LFKITEISPE CAYSTETTKV VVVGNFDEQY KHLTGSAMYV 360 VFGDQCMAAD IVQTGVYRFM VGPHTPGQVD FYLTIDGKTP ISEIRSFTYH VMHANSLGGR 420 FTPSEDEYKK SNLQMQMRLA RLLFATNKKK IAPKLLVEGS KVSNLISASP EKEWTNLWNI 480 LSNSEGSEGV TESLLELVLR NRLQEWLVEM VMEGHKSTGR DDLGQGAIHL CSFLGFSLDF 540 RDSSGWTALH WAAYHGRERM VAALLSAGAN PSLVTDPTPE FPAGLTAADL AARQGYDGLA 600 AYLAEKGLTA HFEAMSLSKD TTQSPSKMKL TKLQSEKFEN LSEQELCLKE SLAAYRNAAD 660 AACNIQAALR ERTLKLQTKA IQLANPEIEA SEIVAAMRIQ HAFRNYNRKK TMRAAARIQS 720 HFRTWKMRRN FINMRRQAIR IQAAYRGHQV RRQYRKVIWS VGIVEKAILR WRKKRKGLRG 780 IASGMPVVMT VDAEAELAST AEEDFFQAGR QQAEDRFNRS VVRVQALFRS YKAQQEYRRM 840 KIAHEEAKLE FSEGQLGAA |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator that binds calmodulin in a calcium-dependent manner in vitro. Binds to the DNA consensus sequence 5'-T[AC]CG[CT]GT[GT][GT][GT][GT]T[GT]CG-3'. {ECO:0000269|PubMed:16192280}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00435 | DAP | Transfer from AT4G16150 | Download |
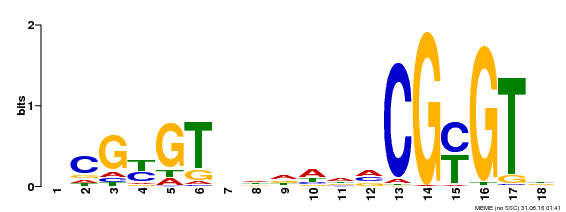
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | LPERR07G12350.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK065480 | 0.0 | AK065480.1 Oryza sativa Japonica Group cDNA clone:J013025F20, full insert sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015646432.1 | 0.0 | calmodulin-binding transcription activator CBT-like isoform X2 | ||||
| Swissprot | Q7XHR2 | 0.0 | CBT_ORYSJ; Calmodulin-binding transcription activator CBT | ||||
| TrEMBL | A0A0D9WYY6 | 0.0 | A0A0D9WYY6_9ORYZ; Uncharacterized protein | ||||
| STRING | LPERR07G12350.2 | 0.0 | (Leersia perrieri) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G16150.1 | 0.0 | calmodulin binding;transcription regulators | ||||



