 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | LPERR08G03480.2 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Leersia
|
||||||||
| Family | MYB_related | ||||||||
| Protein Properties | Length: 770aa MW: 84257.4 Da PI: 7.1479 | ||||||||
| Description | MYB_related family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 47.7 | 3.5e-15 | 73 | 117 | 1 | 47 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
r rWT+ E++++++a k++G W +I +++g ++t+ q++s+ qk+
LPERR08G03480.2 73 RERWTEAEHKRFLEALKLYGRA-WQRIEEHVG-TKTAVQIRSHAQKF 117
78******************88.*********.************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 3.14E-16 | 67 | 123 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 19.147 | 68 | 122 | IPR017930 | Myb domain |
| TIGRFAMs | TIGR01557 | 1.8E-16 | 71 | 120 | IPR006447 | Myb domain, plants |
| SMART | SM00717 | 6.5E-12 | 72 | 120 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 1.6E-12 | 73 | 116 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 8.5E-8 | 73 | 113 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 1.14E-8 | 75 | 118 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009409 | Biological Process | response to cold | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0009733 | Biological Process | response to auxin | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0009739 | Biological Process | response to gibberellin | ||||
| GO:0009751 | Biological Process | response to salicylic acid | ||||
| GO:0009753 | Biological Process | response to jasmonic acid | ||||
| GO:0010243 | Biological Process | response to organonitrogen compound | ||||
| GO:0042754 | Biological Process | negative regulation of circadian rhythm | ||||
| GO:0043496 | Biological Process | regulation of protein homodimerization activity | ||||
| GO:0045892 | Biological Process | negative regulation of transcription, DNA-templated | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0046686 | Biological Process | response to cadmium ion | ||||
| GO:0048574 | Biological Process | long-day photoperiodism, flowering | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 770 aa Download sequence Send to blast |
MTMLLSRRRR GRPWPACGGL GLLRGASQRA RRQRTRLKEV SGLSRLTAIG SIQINSSGEE 60 AMVRKPYTIT KQRERWTEAE HKRFLEALKL YGRAWQRIEE HVGTKTAVQI RSHAQKFFTK 120 LEKEAINNGT SPGQAHDIDI PPPRPKRKPN SPYPRKSCLS SETSTREIPN DKATKSNLTV 180 NSTAQMAGDA ALEKLQRKEI SEKGSCSEVL NLFREAPSAS FSSVNKSSSN HGASRGLEPT 240 KTEIKDVVIL ERDGISNGAG KNVKDINDQE TERLNGIHIS SKSDHSHENC LDTSRQQLKP 300 KATAVEPTYV DWSAAKTSHC QMDRNGATGF PATGTEGSHP DQTSDQTGRT SETMNQCIHP 360 TLSVDPKFDS TATAQPFPHN YAAFAPMMQC HCNQDSYRSF VNMSSTFSSM LVSTLLSNPA 420 IHAAARLAAS YWPAVDGNSA DPNQEDLSES AQGSHAGSPP NMASIVAATV AAASAWWATQ 480 GLLPLFPPPI AFPFVPAPST PFPAADVQRA PEKDMNCPID SAQKEMQETQ KQDNPEAIKV 540 IMSSESDESG KGEVSLHTEL KISPADKADA TPTTGADTSD VFGNKKKQDR SSCGSNTPSS 600 SDHDIEADNA PEKQEKANDK AKQASCSNSS AGDNNHRRFR SSGSTSDSWK EVSEEGRLAF 660 DALFSRERLP QSFSPPQAEE SKEISKEEED EVTTVTVDLN KNATIIDHEL DTVDDPRAFF 720 PNELSNLKLK SRRTGFKPYK RCSVEAKENR VPASDEVGTK RIRLDSETST |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 28 | 35 | RARRQRTR |
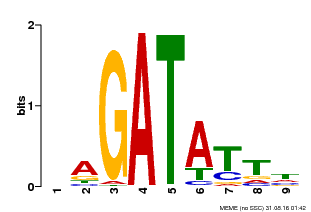
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00654 | PBM | Transfer from LOC_Os08g06110 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | LPERR08G03480.2 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK242771 | 0.0 | AK242771.1 Oryza sativa Japonica Group cDNA, clone: J090053J23, full insert sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015649844.1 | 0.0 | protein LHY isoform X2 | ||||
| Refseq | XP_015649845.1 | 0.0 | protein LHY isoform X2 | ||||
| TrEMBL | A0A0D9X4L6 | 0.0 | A0A0D9X4L6_9ORYZ; Uncharacterized protein | ||||
| STRING | LPERR08G03480.1 | 0.0 | (Leersia perrieri) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G01060.4 | 2e-37 | MYB_related family protein | ||||



