 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | LPERR08G03510.2 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Leersia
|
||||||||
| Family | NAC | ||||||||
| Protein Properties | Length: 690aa MW: 76592.9 Da PI: 4.4737 | ||||||||
| Description | NAC family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | NAM | 166.2 | 1.1e-51 | 9 | 142 | 1 | 128 |
NAM 1 lppGfrFhPtdeelvveyLkkkvegkkleleevikevdiykvePwdLp.k.kvkaeekewyfFskrdkkyatgkrknratksgyWkatgkdkev 92
lp GfrFhPtdeelv +yLk k++g+ ++evi+e+d++k+ePwdLp k +++++ ew+fF+++d+ky++g+r+nrat++gyWkatgkd+ +
LPERR08G03510.2 9 LPLGFRFHPTDEELVRHYLKGKITGQIRGETEVIPEIDVCKCEPWDLPdKsLIRSDDPEWFFFAPKDRKYPNGSRSNRATEAGYWKATGKDRII 102
699************************99999***************96446677888************************************ PP
NAM 93 lsk....kgelvglkktLvfykgrapkgektdWvmheyrl 128
+sk k++ +g+kktLvf++grapkge+t W+mheyr
LPERR08G03510.2 103 KSKgdkkKQHMIGMKKTLVFHRGRAPKGERTGWIMHEYRT 142
**977655666***************************96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF101941 | 6.15E-58 | 3 | 163 | IPR003441 | NAC domain |
| PROSITE profile | PS51005 | 56.992 | 9 | 163 | IPR003441 | NAC domain |
| Pfam | PF02365 | 1.2E-27 | 10 | 141 | IPR003441 | NAC domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0045892 | Biological Process | negative regulation of transcription, DNA-templated | ||||
| GO:0071470 | Biological Process | cellular response to osmotic stress | ||||
| GO:1900426 | Biological Process | positive regulation of defense response to bacterium | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005789 | Cellular Component | endoplasmic reticulum membrane | ||||
| GO:0005886 | Cellular Component | plasma membrane | ||||
| GO:0016021 | Cellular Component | integral component of membrane | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0005516 | Molecular Function | calmodulin binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 690 aa Download sequence Send to blast |
MTVMELKKLP LGFRFHPTDE ELVRHYLKGK ITGQIRGETE VIPEIDVCKC EPWDLPDKSL 60 IRSDDPEWFF FAPKDRKYPN GSRSNRATEA GYWKATGKDR IIKSKGDKKK QHMIGMKKTL 120 VFHRGRAPKG ERTGWIMHEY RTTEPQFESG EQGGYVLYRL FRKQEEKIER PSLDEMDRSG 180 YSPTPSRSTP DNMEPNEDGN TPLNKESPES ALHESPIDLP ALTEAPAAPI TRWLADTADN 240 VTTNEANISH MPFHGLDGGA KASPSVGAST QLIHSRKNIY DNDELATVSA PMLPLQDFGE 300 FPLGAIGNFD GNMKPRDPVE EFLNQTIADP DEHSSTTSKA QYDSDTGIVL TEFENPGMMQ 360 GGFMDDPSGL ENLSFWPDDT NPQLSALYEN ATLLPYDISA DQDVLSMDSG AESLQDLFNS 420 LEDSSTKINT WNNEPVFQGT GIEIYEQLQS NDIFVNQGTA RRRLRLQQSE SLSPDFEDEE 480 SGIVVTSKYV NEVAEESTAE KDMPSDGDDA ESTGITILRR RQAPTASSDD EEAESTGITI 540 LRRRHAPTAS SGSFTQQGAA VRRVRLQCDL NAAPCSSGDG SSSCIIDESE SECTMEKAEI 600 EEHTRTTLAE GVDLSGPEHD ASAVVADDKS VLRQRKTAEG SDKENKQDYC VLDSHVRAPV 660 KKRGFPAYII WLVLSLALVL LIFVGIYGWV |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ut4_A | 3e-45 | 9 | 164 | 17 | 166 | NO APICAL MERISTEM PROTEIN |
| 1ut4_B | 3e-45 | 9 | 164 | 17 | 166 | NO APICAL MERISTEM PROTEIN |
| 1ut7_A | 3e-45 | 9 | 164 | 17 | 166 | NO APICAL MERISTEM PROTEIN |
| 1ut7_B | 3e-45 | 9 | 164 | 17 | 166 | NO APICAL MERISTEM PROTEIN |
| 3swm_A | 3e-45 | 9 | 164 | 20 | 169 | NAC domain-containing protein 19 |
| 3swm_B | 3e-45 | 9 | 164 | 20 | 169 | NAC domain-containing protein 19 |
| 3swm_C | 3e-45 | 9 | 164 | 20 | 169 | NAC domain-containing protein 19 |
| 3swm_D | 3e-45 | 9 | 164 | 20 | 169 | NAC domain-containing protein 19 |
| 3swp_A | 3e-45 | 9 | 164 | 20 | 169 | NAC domain-containing protein 19 |
| 3swp_B | 3e-45 | 9 | 164 | 20 | 169 | NAC domain-containing protein 19 |
| 3swp_C | 3e-45 | 9 | 164 | 20 | 169 | NAC domain-containing protein 19 |
| 3swp_D | 3e-45 | 9 | 164 | 20 | 169 | NAC domain-containing protein 19 |
| 4dul_A | 3e-45 | 9 | 164 | 17 | 166 | NAC domain-containing protein 19 |
| 4dul_B | 3e-45 | 9 | 164 | 17 | 166 | NAC domain-containing protein 19 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00108 | PBM | Transfer from AT4G35580 | Download |
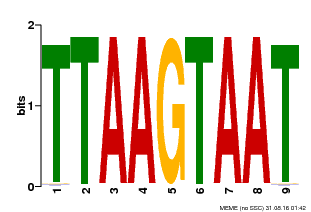
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | LPERR08G03510.2 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | EU846994 | 0.0 | EU846994.1 Oryza sativa Japonica Group clone KCS281G09 NAC2 protein mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015650289.1 | 0.0 | NAC domain-containing protein 14 isoform X2 | ||||
| TrEMBL | A0A0D9X4M0 | 0.0 | A0A0D9X4M0_9ORYZ; Uncharacterized protein | ||||
| STRING | LPERR08G03510.1 | 0.0 | (Leersia perrieri) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G35580.1 | 1e-70 | NAC transcription factor-like 9 | ||||



