 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | LPERR09G10870.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Leersia
|
||||||||
| Family | EIL | ||||||||
| Protein Properties | Length: 616aa MW: 68816.9 Da PI: 6.3351 | ||||||||
| Description | EIL family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | EIN3 | 419.7 | 3.2e-128 | 38 | 417 | 1 | 353 |
XXXXXXXXXXXXXXXXXXXXXXX..XXXXX.XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX CS
EIN3 1 eelkkrmwkdqmllkrlkerkkqlledkeaatgakksnksneqarrkkmsraQDgiLkYMlkemevcnaqGfvYgiipekgkpvegasdsLraW 94
++l +rmwkd+++l+r+ker+ +l +a+++++k + +++qa rkkmsra DgiLkYMlk mevc a+GfvYgiip+kgkpv+gasd++raW
LPERR09G10870.1 38 DDLARRMWKDRVKLRRIKERQHKLA-LVQAELDKSKPKLISDQAMRKKMSRAHDGILKYMLKLMEVCHARGFVYGIIPDKGKPVSGASDNIRAW 130
699******************9966.55566*************************************************************** PP
XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX....XX----STTS-HHHHHHHHHHHSSSSSS-TTS--TTT--HHHH---S--HHHHHHT--T CS
EIN3 95 WkekvefdrngpaaiskyqaknlilsgesslqtersseshslselqDTtlgSLLsalmqhcdppqrrfplekgvepPWWPtGkelwwgelglsk 188
Wkekv+fd+ngpaai+ky+++nl++ ++ s+ + +++hsl++lqD+tlgSLLs+lmq c+ppqr+fpl+kg++pPWWP+G+e+ww +lgl+
LPERR09G10870.1 131 WKEKVKFDKNGPAAIAKYESENLASVDAPSS---GIKSQHSLMDLQDATLGSLLSSLMQNCNPPQRKFPLDKGTPPPWWPSGNEDWWIALGLPV 221
************************9988888...88********************************************************** PP
T--.-----GGG--HHHHHHHHHHHHHHTGGGHHHHHHTTTTSSSSTTT--SHHHHHHHHHHTTTTT-S--XXXXXX.....XXXXXXXXXXXX CS
EIN3 189 dqgtppykkphdlkkawkvsvLtavikhmsptieeirelerqskylqdkmsakesfallsvlnqeekecatvsahss.....slrkqspkvtls 277
q ppykkphdlkk+wkv+vLt vikhmsp++++ir+++r+sk+lqdkm+akes ++l vl++ee++++ + +s + ++ tls
LPERR09G10870.1 222 GQI-PPYKKPHDLKKVWKVGVLTGVIKHMSPNFDKIRNHVRKSKCLQDKMTAKESLIWLGVLQREERLIHCIDNGMSevtqhLALEYRNGDTLS 314
999.9****************************************************************9888885535554333489999*** PP
XXXXXXXXXXXXXX.XXXXXXXXXX................................XXXXXXXXXXXXXXXXXXXXX......XXXXXXX.XX CS
EIN3 278 ceqkedvegkkeskikhvqavktta................................gfpvvrkrkkkpsesakvsskevsrtcqssqfrgset 339
+++++dv+g ++ ++++ +++++ g++ krk kp ++ +v ++ev q++ +++
LPERR09G10870.1 315 SSNEYDVDGFEDAPLSTSSRDDEQDlspaaqlseepaptrreranvkrpnqvvpkkaGTKEPPKRK-KPCHNVTVIEHEV----QRADDAPENS 403
********8887777988888888899****************************99999999999.6666666666655....4444445678 PP
XXXXXXXXXXXXXX CS
EIN3 340 elifadknsisqne 353
+ +++d+n+++q e
LPERR09G10870.1 404 RSMIPDMNRLDQVE 417
99999*99999876 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF04873 | 7.8E-123 | 38 | 286 | No hit | No description |
| Gene3D | G3DSA:1.10.3180.10 | 3.0E-68 | 160 | 291 | IPR023278 | Ethylene insensitive 3-like protein, DNA-binding domain |
| SuperFamily | SSF116768 | 6.02E-60 | 166 | 289 | IPR023278 | Ethylene insensitive 3-like protein, DNA-binding domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0042762 | Biological Process | regulation of sulfur metabolic process | ||||
| GO:0071281 | Biological Process | cellular response to iron ion | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 616 aa Download sequence Send to blast |
MDHLAIIATE LGDSLDFEVD GIQNLTENDV SDEEIEVDDL ARRMWKDRVK LRRIKERQHK 60 LALVQAELDK SKPKLISDQA MRKKMSRAHD GILKYMLKLM EVCHARGFVY GIIPDKGKPV 120 SGASDNIRAW WKEKVKFDKN GPAAIAKYES ENLASVDAPS SGIKSQHSLM DLQDATLGSL 180 LSSLMQNCNP PQRKFPLDKG TPPPWWPSGN EDWWIALGLP VGQIPPYKKP HDLKKVWKVG 240 VLTGVIKHMS PNFDKIRNHV RKSKCLQDKM TAKESLIWLG VLQREERLIH CIDNGMSEVT 300 QHLALEYRNG DTLSSSNEYD VDGFEDAPLS TSSRDDEQDL SPAAQLSEEP APTRRERANV 360 KRPNQVVPKK AGTKEPPKRK KPCHNVTVIE HEVQRADDAP ENSRSMIPDM NRLDQVEMPG 420 MANQITSFNK EGNTSEALQH RGNTQELAHL PVTNFNHYLS AQATDATPVS ICVGGQSVPY 480 EISDNSRPKT GNIFPLDSDS GFNNLPSSYQ TIPPKQSLPL SMMDHHVVPM GIRTPADNSP 540 YVDQIIGSGN STSVPGDMQL IDYPFYGEQD KFAGSLGCDT SWCLFLSSIL SLGYHLNLSM 600 EGYTFFVLPS NINLDS |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wij_A | 5e-63 | 166 | 290 | 9 | 133 | ETHYLENE-INSENSITIVE3-like 3 protein |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor that may be involved in the ethylene response pathway. {ECO:0000269|PubMed:9215635, ECO:0000269|PubMed:9851977}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00649 | PBM | Transfer from LOC_Os09g31400 | Download |
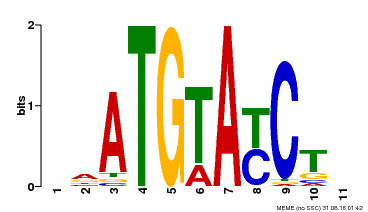
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | LPERR09G10870.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AC108758 | 0.0 | AC108758.2 Oryza sativa (japonica cultivar-group) chromosome 9 BAC clone OSJNBa0066E19, complete sequence. | |||
| GenBank | AK065979 | 0.0 | AK065979.1 Oryza sativa Japonica Group cDNA clone:J013047J08, full insert sequence. | |||
| GenBank | AP014965 | 0.0 | AP014965.1 Oryza sativa Japonica Group DNA, chromosome 9, cultivar: Nipponbare, complete sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015612094.1 | 0.0 | ETHYLENE INSENSITIVE 3-like 3 protein | ||||
| Refseq | XP_015612095.1 | 0.0 | ETHYLENE INSENSITIVE 3-like 3 protein | ||||
| Swissprot | O23116 | 1e-142 | EIL3_ARATH; ETHYLENE INSENSITIVE 3-like 3 protein | ||||
| TrEMBL | A0A0D9XF38 | 0.0 | A0A0D9XF38_9ORYZ; Uncharacterized protein | ||||
| STRING | LPERR09G10870.1 | 0.0 | (Leersia perrieri) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP3720 | 37 | 79 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G73730.1 | 1e-145 | ETHYLENE-INSENSITIVE3-like 3 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



