 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | LPERR09G11330.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Leersia
|
||||||||
| Family | NAC | ||||||||
| Protein Properties | Length: 346aa MW: 38470.9 Da PI: 9.2815 | ||||||||
| Description | NAC family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | NAM | 180.4 | 4.8e-56 | 17 | 147 | 1 | 128 |
NAM 1 lppGfrFhPtdeelvveyLkkkvegkkleleevikevdiykvePwdLpkkvkaeekewyfFskrdkkyatgkrknratksgyWkatgkdkevls 94
lppGfrFhPtdeel+++yL++k+++ ++++ ++i+evd++k+ePwdLp+k+k +ekewyfFs rd+ky+tg r+nrat++gyWk+tgkdke+++
LPERR09G11330.1 17 LPPGFRFHPTDEELITYYLRQKIADGSFTA-RAIAEVDLNKCEPWDLPEKAKMGEKEWYFFSLRDRKYPTGVRTNRATNAGYWKTTGKDKEIFT 109
79*************************999.89***************99999****************************************9 PP
NAM 95 k....kgelvglkktLvfykgrapkgektdWvmheyrl 128
+ elvg+kktLvfykgrap+gekt+Wvmheyrl
LPERR09G11330.1 110 GqppaTPELVGMKKTLVFYKGRAPRGEKTNWVMHEYRL 147
766666677***************************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF101941 | 3.01E-64 | 14 | 168 | IPR003441 | NAC domain |
| PROSITE profile | PS51005 | 60.935 | 17 | 168 | IPR003441 | NAC domain |
| Pfam | PF02365 | 2.3E-29 | 18 | 147 | IPR003441 | NAC domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 346 aa Download sequence Send to blast |
MGMEGSGGGS EKKEESLPPG FRFHPTDEEL ITYYLRQKIA DGSFTARAIA EVDLNKCEPW 60 DLPEKAKMGE KEWYFFSLRD RKYPTGVRTN RATNAGYWKT TGKDKEIFTG QPPATPELVG 120 MKKTLVFYKG RAPRGEKTNW VMHEYRLHSK SIPKSNKDEW VVCRVFAKTA GVKKYPSNNV 180 HSRSHHPYTL DMVPPLLPAL LQQDPFGRGH HPYMTPVDMA ELSRFARGTP GLHPHIQPHP 240 GYINPAGPFT LSGLNLNLGS SPAMPPPPPQ SILQTMSMSM PMNQPSTANQ VMATEQMVPG 300 LGNGVIPPGA DGGFTTDAVV GGTSIRYQNL DVEQLVERYW PGSYQM |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ut4_A | 1e-55 | 16 | 174 | 16 | 171 | NO APICAL MERISTEM PROTEIN |
| 1ut4_B | 1e-55 | 16 | 174 | 16 | 171 | NO APICAL MERISTEM PROTEIN |
| 1ut7_A | 1e-55 | 16 | 174 | 16 | 171 | NO APICAL MERISTEM PROTEIN |
| 1ut7_B | 1e-55 | 16 | 174 | 16 | 171 | NO APICAL MERISTEM PROTEIN |
| 3swm_A | 2e-55 | 16 | 174 | 19 | 174 | NAC domain-containing protein 19 |
| 3swm_B | 2e-55 | 16 | 174 | 19 | 174 | NAC domain-containing protein 19 |
| 3swm_C | 2e-55 | 16 | 174 | 19 | 174 | NAC domain-containing protein 19 |
| 3swm_D | 2e-55 | 16 | 174 | 19 | 174 | NAC domain-containing protein 19 |
| 3swp_A | 2e-55 | 16 | 174 | 19 | 174 | NAC domain-containing protein 19 |
| 3swp_B | 2e-55 | 16 | 174 | 19 | 174 | NAC domain-containing protein 19 |
| 3swp_C | 2e-55 | 16 | 174 | 19 | 174 | NAC domain-containing protein 19 |
| 3swp_D | 2e-55 | 16 | 174 | 19 | 174 | NAC domain-containing protein 19 |
| 4dul_A | 1e-55 | 16 | 174 | 16 | 171 | NAC domain-containing protein 19 |
| 4dul_B | 1e-55 | 16 | 174 | 16 | 171 | NAC domain-containing protein 19 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Binds to the promoter regions of genes involved in chlorophyll catabolic processes, such as NYC1, SGR1, SGR2 and PAO. {ECO:0000269|PubMed:27021284}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00277 | DAP | Transfer from AT2G24430 | Download |
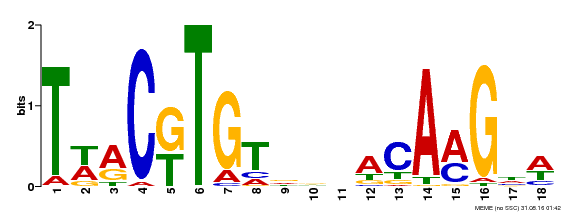
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | LPERR09G11330.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Repressed by the microRNA miR164. {ECO:0000269|PubMed:15294871, ECO:0000269|PubMed:17098808}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK102511 | 0.0 | AK102511.1 Oryza sativa Japonica Group cDNA clone:J033095I08, full insert sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006660793.1 | 0.0 | PREDICTED: NAC domain-containing protein 92-like | ||||
| Swissprot | Q9FLJ2 | 4e-84 | NC100_ARATH; NAC domain-containing protein 100 | ||||
| TrEMBL | A0A0D9XF88 | 0.0 | A0A0D9XF88_9ORYZ; Uncharacterized protein | ||||
| STRING | LPERR09G11330.1 | 0.0 | (Leersia perrieri) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP9361 | 38 | 46 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G24430.2 | 2e-90 | NAC domain containing protein 38 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



