 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | LPERR09G11920.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Leersia
|
||||||||
| Family | SBP | ||||||||
| Protein Properties | Length: 450aa MW: 47760 Da PI: 8.1692 | ||||||||
| Description | SBP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SBP | 129.2 | 1.5e-40 | 111 | 187 | 2 | 78 |
-SSTT-----TT--HHHHHTT--HHHHT-S-EEETTEEEEE-TTTSSEEETTT--SS--S-STTTT-------S--- CS
SBP 2 CqvegCeadlseakeyhrrhkvCevhskapvvlvsgleqrfCqqCsrfhelsefDeekrsCrrrLakhnerrrkkqa 78
C v+gC+adls+ ++yhrrhkvCe hsk+pvv+vsg+e rfCqqCsrfh l efDe krsCr+rL++hn+rrrk+qa
LPERR09G11920.1 111 CAVDGCKADLSKYRDYHRRHKVCEPHSKTPVVVVSGREMRFCQQCSRFHLLGEFDEIKRSCRKRLDGHNRRRRKPQA 187
**************************************************************************986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.1100.10 | 5.8E-35 | 104 | 172 | IPR004333 | Transcription factor, SBP-box |
| PROSITE profile | PS51141 | 31.752 | 108 | 185 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF103612 | 2.62E-38 | 109 | 190 | IPR004333 | Transcription factor, SBP-box |
| Pfam | PF03110 | 2.3E-31 | 111 | 184 | IPR004333 | Transcription factor, SBP-box |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0048653 | Biological Process | anther development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 450 aa Download sequence Send to blast |
MDWDLKMPVS WDLTDLDHDA VPATPATNTT TALGIASAAV APAGRPECSV DLKLGGLGEF 60 GAAAKGPAVV PPSNTSPASP AATAVARVPT AAASPARRPR GVSGAGQCPS CAVDGCKADL 120 SKYRDYHRRH KVCEPHSKTP VVVVSGREMR FCQQCSRFHL LGEFDEIKRS CRKRLDGHNR 180 RRRKPQADSM SASFMTSQQG TRFASFTPPR PEPSWSGIIK SEETPYYSHH HHHHHHQIMS 240 SGKQHFVGSP SRTTTTTTTA TFSSSSSKEA AGRRFPFLHD GDQISLLKTT TAVVSPVPAT 300 AIESSSSNNN KIFSDGQLGH THHQILDSDC ALSLLSSPAN SSTVDVAGRI NLHHHPPHHP 360 SPAAIPIAQS LVPNLQQQFG GGGGSSPWFA SGGGGGGFNC PTSVESEQQQ VNTAVVPGSN 420 ENEMNYHGIF HVAGEGSSDG TSPSIPFSWQ |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ul4_A | 8e-29 | 101 | 184 | 1 | 84 | squamosa promoter binding protein-like 4 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 167 | 184 | KRSCRKRLDGHNRRRRKP |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Trans-acting factor that binds specifically to the consensus nucleotide sequence 5'-TNCGTACAA-3' (By similarity). May be involved in panicle development. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00555 | DAP | Transfer from AT5G50570 | Download |
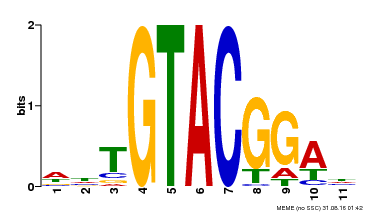
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | LPERR09G11920.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Negatively regulated by microRNAs miR156b and miR156h. {ECO:0000305|PubMed:16861571}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK109469 | 4e-68 | AK109469.1 Oryza sativa Japonica Group cDNA clone:001-128-D02, full insert sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015610874.1 | 1e-163 | squamosa promoter-binding-like protein 18 isoform X2 | ||||
| Swissprot | Q0J0K1 | 1e-164 | SPL18_ORYSJ; Squamosa promoter-binding-like protein 18 | ||||
| TrEMBL | A0A0D9XFE9 | 0.0 | A0A0D9XFE9_9ORYZ; Uncharacterized protein | ||||
| STRING | LPERR09G11920.1 | 0.0 | (Leersia perrieri) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP1755 | 37 | 95 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G50670.1 | 2e-40 | SBP family protein | ||||



