 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | LPERR12G07400.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Leersia
|
||||||||
| Family | C2H2 | ||||||||
| Protein Properties | Length: 1252aa MW: 139814 Da PI: 7.8042 | ||||||||
| Description | C2H2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-C2H2 | 13 | 0.00032 | 1133 | 1156 | 1 | 22 |
EEET..TTTEEESSHHHHHHHHHH CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirt 22
++C C++sFs++ +L H r
LPERR12G07400.1 1133 FPCDieGCDMSFSTQQDLLLHKRD 1156
789999***************985 PP
| |||||||
| 2 | zf-C2H2 | 12.3 | 0.00053 | 1216 | 1242 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHH..T CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirt..H 23
y+C+ Cg++F+ s++ rH r+ H
LPERR12G07400.1 1216 YECKepGCGQTFRFVSDFSRHKRKtgH 1242
89********************99666 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51184 | 34.061 | 64 | 233 | IPR003347 | JmjC domain |
| SMART | SM00558 | 2.7E-44 | 64 | 233 | IPR003347 | JmjC domain |
| SuperFamily | SSF51197 | 1.02E-22 | 80 | 230 | No hit | No description |
| Pfam | PF02373 | 4.6E-35 | 97 | 216 | IPR003347 | JmjC domain |
| SMART | SM00355 | 9.7 | 1133 | 1155 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 10.907 | 1156 | 1185 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 15 | 1156 | 1180 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 1158 | 1180 | IPR007087 | Zinc finger, C2H2 |
| SuperFamily | SSF57667 | 2.66E-14 | 1167 | 1222 | No hit | No description |
| Gene3D | G3DSA:3.30.160.60 | 1.6E-10 | 1183 | 1210 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE profile | PS50157 | 11.323 | 1186 | 1215 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.0018 | 1186 | 1210 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 1188 | 1210 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 9.6E-10 | 1211 | 1239 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE profile | PS50157 | 11.926 | 1216 | 1247 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.15 | 1216 | 1242 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 1218 | 1242 | IPR007087 | Zinc finger, C2H2 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009741 | Biological Process | response to brassinosteroid | ||||
| GO:0009826 | Biological Process | unidimensional cell growth | ||||
| GO:0010228 | Biological Process | vegetative to reproductive phase transition of meristem | ||||
| GO:0033169 | Biological Process | histone H3-K9 demethylation | ||||
| GO:0035067 | Biological Process | negative regulation of histone acetylation | ||||
| GO:0048366 | Biological Process | leaf development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003676 | Molecular Function | nucleic acid binding | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1252 aa Download sequence Send to blast |
MAAEFEPPRH ASPPKNATSL QIEALFWAAC ASKPFSVEYG NDMPGSGFAS PEELGQKLGA 60 NANDVGETEW NMRVAPRARG SLLRAMGRDV AGVTSPMLYV AMLYSWFAWH VEDHELHSLN 120 FLHFGKPKTW YGVPRDAMLA FEETVRVHGY ADDLNAIMAF QTLNEKTTVL SPEVLLSAGV 180 PCCRLVQKAG EFVITFPGAY HSGFSHGFNC GEASNIATPH WLQVAKEAAI RRASTNCGPM 240 VSHYQLLYEL ALSLRPREPN NFHFVRSSRL RDKNKNEGDI MVKENFVGSV TENNNLLSVL 300 LDKNSCIIVP KIAFPISSFR MALESEVTAK QKFTAGPSSI SQQGAENMAV DHVSVDKASE 360 IQDMSGSIYA CDTSFVACSS RKLYETKYGK KDAASLCLST SEIQSRGIDN ARLHPAGGIL 420 DQGRLPCVQC GILSFACVAI IQPREAAVQF IMSKECISSS ANHGEIGASD DTSNWINRHH 480 EIGPPPGIAS ETEDNVKHTT SLAHVSDQCE QLYGNKTDGG TSALGLLASA YDSSDSDEET 540 AEDVSKHREK NDSVNENTNT RILETSSSCS SMVQCQKKNS HLHGKCEARD TSLMEPVNHN 600 GRTISQCSRD TDFGHFIELG KSGTHCSGYL DLVDDLTTSV LKPSSDTYVS ADKASMDPDV 660 LTMLKYNKES CRMHVFCLEH ALETWTQLQK IGGANIMLLC HPEYPRAESA AKVIAEELGV 720 KHDWKDVNFK EATEEDIRKI QLALQDEDAE PTGSDWAVKM GINIYYSAKQ SKSPLYSKQI 780 PYNSIIYKAF GQENPDSLTD YGGQKSGSAK KKVAGWWCGK VWMSNQVHPL LARQREEQNG 840 STGYSKITFA AISHAKVQDE PSTRCNTLIN RSLSKRTSRR KGMESAEKSK EKKKKTTASN 900 EASIHYNGPG INSEVISDQL ENSDDHDKYD KRDEIEEETN PQIYQQHKLQ NVTRKSVSRK 960 RKDEKRKDGF HELYGKDNDV DYLVNNTNMG SGDDATLGNS DNTQQQSLDP VKVKSGGKLQ 1020 GNKRKSSKCK SSDDLLNEDN KLQKMNKKSS SKKQKNDKIN RQFQGQNEDD HLDHLLDVEI 1080 RDVMPENEDK ITEDKIEDAE VKSRGKLQSG KSKTSKRQAR DSLRTGNNKL AKFPCDIEGC 1140 DMSFSTQQDL LLHKRDICPV KGCKKKFFCH KYLLQHRKVH IDERPLKCMW KGCKKAFKWP 1200 WARTEHMRVH TGVRPYECKE PGCGQTFRFV SDFSRHKRKT GHSSDKRRKN ST |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6a57_A | 2e-56 | 1130 | 1249 | 20 | 139 | Lysine-specific demethylase REF6 |
| 6a58_A | 2e-56 | 1130 | 1249 | 20 | 139 | Lysine-specific demethylase REF6 |
| 6a59_A | 2e-56 | 1130 | 1249 | 20 | 139 | Lysine-specific demethylase REF6 |
| 6ip0_A | 2e-53 | 22 | 254 | 125 | 353 | Transcription factor jumonji (Jmj) family protein |
| 6ip4_A | 2e-53 | 22 | 254 | 125 | 353 | Arabidopsis JMJ13 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 878 | 894 | RRKGMESAEKSKEKKKK |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00608 | ChIP-seq | Transfer from AT3G48430 | Download |
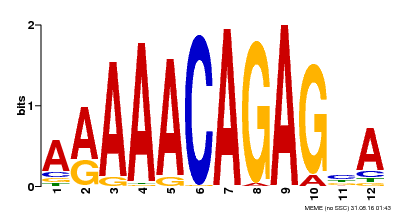
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | LPERR12G07400.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK065251 | 0.0 | AK065251.1 Oryza sativa Japonica Group cDNA clone:J013002J08, full insert sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015620626.1 | 0.0 | lysine-specific demethylase REF6 | ||||
| TrEMBL | A0A0D9XYG9 | 0.0 | A0A0D9XYG9_9ORYZ; Uncharacterized protein | ||||
| STRING | LPERR12G07400.1 | 0.0 | (Leersia perrieri) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP6718 | 30 | 35 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G48430.1 | 1e-126 | relative of early flowering 6 | ||||



