 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | LPERR12G11080.2 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Leersia
|
||||||||
| Family | WRKY | ||||||||
| Protein Properties | Length: 368aa MW: 40510.4 Da PI: 7.4532 | ||||||||
| Description | WRKY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | WRKY | 100.7 | 8.9e-32 | 98 | 154 | 2 | 59 |
--SS-EEEEEEE--TT-SS-EEEEEE-STT---EEEEEE-SSSTTEEEEEEES--SS- CS
WRKY 2 dDgynWrKYGqKevkgsefprsYYrCtsagCpvkkkversaedpkvveitYegeHnhe 59
dDgynWrKYGqK+vkgs++prsYY+Ct+++Cpvkkkve+++ d+++ ei+Y+g+Hnh+
LPERR12G11080.2 98 DDGYNWRKYGQKVVKGSDCPRSYYKCTHPSCPVKKKVEHAE-DGQISEIIYKGKHNHQ 154
8**************************************99.***************8 PP
| |||||||
| 2 | WRKY | 108.6 | 3e-34 | 254 | 312 | 1 | 59 |
---SS-EEEEEEE--TT-SS-EEEEEE-STT---EEEEEE-SSSTTEEEEEEES--SS- CS
WRKY 1 ldDgynWrKYGqKevkgsefprsYYrCtsagCpvkkkversaedpkvveitYegeHnhe 59
ldDgy+WrKYGqK+vkg+++prsYY+Ct+agC+v+k++er+++dpk+v++tYeg+Hnhe
LPERR12G11080.2 254 LDDGYRWRKYGQKVVKGNPHPRSYYKCTFAGCNVRKHIERASSDPKAVITTYEGKHNHE 312
59********************************************************8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:2.20.25.80 | 1.9E-28 | 94 | 156 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 3.14E-25 | 96 | 156 | IPR003657 | WRKY domain |
| SMART | SM00774 | 1.1E-33 | 97 | 155 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 1.8E-24 | 98 | 154 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 23.192 | 98 | 156 | IPR003657 | WRKY domain |
| Gene3D | G3DSA:2.20.25.80 | 5.0E-37 | 240 | 314 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 1.83E-29 | 246 | 314 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 37.413 | 249 | 314 | IPR003657 | WRKY domain |
| SMART | SM00774 | 7.0E-40 | 254 | 313 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 2.6E-26 | 255 | 312 | IPR003657 | WRKY domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 368 aa Download sequence Send to blast |
MLFSPAMGGF GMSHREALAQ VTAQASHSPL RMFDHTEQPS FSAAPTSSET MHHMSAGVNM 60 TGISDMVTGP TNNDNVGFQS AEVSQRYQVN APVDKPADDG YNWRKYGQKV VKGSDCPRSY 120 YKCTHPSCPV KKKVEHAEDG QISEIIYKGK HNHQRPPNKR AKDGSSSAAE QNEQSNDTTS 180 GMSGVKRDQE AIYGMSEQLS GLSDGDDMDD GESRPHEADD KEGDNKKRNI QISSQRTSAE 240 AKIIVQTTSE VDLLDDGYRW RKYGQKVVKG NPHPRSYYKC TFAGCNVRKH IERASSDPKA 300 VITTYEGKHN HEPPVGRGNN QNAGNAAPSS SSQQNVHNMS SNQASLTRAD YNNINQRPIG 360 VLQFKSEE |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wj2_A | 1e-39 | 98 | 314 | 8 | 77 | Probable WRKY transcription factor 4 |
| 2lex_A | 1e-39 | 98 | 314 | 8 | 77 | Probable WRKY transcription factor 4 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor. Interacts specifically with the W box (5'-(T)TGAC[CT]-3'), a frequently occurring elicitor-responsive cis-acting element (By similarity). {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00260 | DAP | Transfer from AT2G03340 | Download |
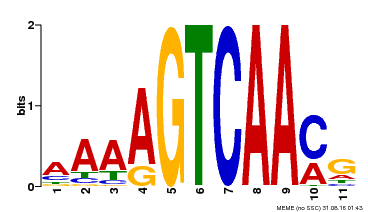
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | LPERR12G11080.2 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By salicylic acid and during leaf senescence. {ECO:0000269|PubMed:11449049, ECO:0000269|PubMed:11722756}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AL731759 | 0.0 | AL731759.2 Oryza sativa chromosome 12, . BAC OSJNBa0009K11 of library OSJNBa from chromosome 12 of cultivar Nipponbare of ssp. japonica of Oryza sativa (rice), complete sequence. | |||
| GenBank | AP014968 | 0.0 | AP014968.1 Oryza sativa Japonica Group DNA, chromosome 12, cultivar: Nipponbare, complete sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015620542.1 | 0.0 | probable WRKY transcription factor 3 | ||||
| Swissprot | Q9ZQ70 | 1e-107 | WRKY3_ARATH; Probable WRKY transcription factor 3 | ||||
| TrEMBL | A0A0D9XZP3 | 0.0 | A0A0D9XZP3_9ORYZ; Uncharacterized protein | ||||
| STRING | LPERR12G11080.2 | 0.0 | (Leersia perrieri) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP1345 | 38 | 122 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G03340.1 | 1e-103 | WRKY DNA-binding protein 3 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



