 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Lj0g3v0140239.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Loteae; Lotus
|
||||||||
| Family | ARF | ||||||||
| Protein Properties | Length: 614aa MW: 68665.1 Da PI: 6.1054 | ||||||||
| Description | ARF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | B3 | 70.6 | 2e-22 | 69 | 169 | 1 | 98 |
EEEE-..-HHHHTT-EE--HHH.HTT.......---..--SEEEEEETTS-EEEEEE..EEETTEEEE-TTHHHHHHHHT--TT-EEEEEE-SS CS
B3 1 ffkvltpsdvlksgrlvlpkkfaeeh.......ggkkeesktltledesgrsWevkliyrkksgryvltkGWkeFvkangLkegDfvvFkldgr 87
f+k+lt sd++++g +++ +++a+++ ++++ + +l+ +d++g++W +++i+r++++r++lt+GW+ Fv++++L +gD ++F +
Lj0g3v0140239.1 69 FCKTLTASDTSTHGGFSVLRRHADDClppldmtQQPPWQ--ELVATDLHGNEWHFRHIFRGQPRRHLLTTGWSVFVSSKKLVAGDAFIFL--RG 158
99****************************975444444..8************************************************..44 PP
SEE..EEEEE- CS
B3 88 sefelvvkvfr 98
+++el+v+v+r
Lj0g3v0140239.1 159 ENGELRVGVRR 169
9999*****97 PP
| |||||||
| 2 | Auxin_resp | 111 | 1.2e-36 | 195 | 277 | 1 | 83 |
Auxin_resp 1 aahaastksvFevvYnPrastseFvvkvekvekalkvkvsvGmRfkmafetedsserrlsGtvvgvsdldpvrWpnSkWrsLk 83
a+ha++t++ F+v+Y+Pr+s+seF+v+v+k+++a ++k+svGmRfkm+fe+e+++err+sGt+vgv+ +++ W +S+WrsLk
Lj0g3v0140239.1 195 ASHAIATGTLFSVFYKPRTSRSEFIVSVNKYLEARNHKLSVGMRFKMRFEGEEVPERRFSGTIVGVEYNNSSVWADSEWRSLK 277
789******************************************************************************96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF101936 | 9.55E-44 | 59 | 198 | IPR015300 | DNA-binding pseudobarrel domain |
| Gene3D | G3DSA:2.40.330.10 | 1.2E-40 | 66 | 184 | IPR015300 | DNA-binding pseudobarrel domain |
| CDD | cd10017 | 6.93E-21 | 67 | 169 | No hit | No description |
| PROSITE profile | PS50863 | 11.251 | 69 | 171 | IPR003340 | B3 DNA binding domain |
| Pfam | PF02362 | 3.7E-20 | 69 | 169 | IPR003340 | B3 DNA binding domain |
| SMART | SM01019 | 1.2E-21 | 69 | 171 | IPR003340 | B3 DNA binding domain |
| Pfam | PF06507 | 7.9E-35 | 195 | 277 | IPR010525 | Auxin response factor |
| Pfam | PF02309 | 1.0E-11 | 487 | 580 | IPR033389 | AUX/IAA domain |
| PROSITE profile | PS51745 | 26.102 | 490 | 583 | IPR000270 | PB1 domain |
| SuperFamily | SSF54277 | 4.32E-11 | 492 | 567 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009733 | Biological Process | response to auxin | ||||
| GO:0010150 | Biological Process | leaf senescence | ||||
| GO:0045892 | Biological Process | negative regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0042802 | Molecular Function | identical protein binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 614 aa Download sequence Send to blast |
MNQGLEHQMP SFNLPCKILC KVVNIHLRAE PETDEVYAQI TLLPETDQSE VASPDEPLPE 60 PTRCTVHSFC KTLTASDTST HGGFSVLRRH ADDCLPPLDM TQQPPWQELV ATDLHGNEWH 120 FRHIFRGQPR RHLLTTGWSV FVSSKKLVAG DAFIFLRGEN GELRVGVRRL MRQQSNMPSS 180 VISSHSMHLG VLATASHAIA TGTLFSVFYK PRTSRSEFIV SVNKYLEARN HKLSVGMRFK 240 MRFEGEEVPE RRFSGTIVGV EYNNSSVWAD SEWRSLKVQW DEPSSILRPD RVSPWELEPL 300 VSTPPANSQP TPRNKRSRPP VLPSTIPDSS QGVWKSPVDS PPFSYRDPQH GRDLYASPKF 360 NSTATSFFCF GGNNSASNKS TYWSTRMENS TETFSPIALK ESGEKRQGTG NGCRLFGIQL 420 LENSNAEESL PTVTLSGRLG DDQSIPSLDA ESDQHSEPSN VNRSDIPSVS CDAEKSCLRS 480 PQESQSRQIR SCTKVHMQGM AVGRAVDLTQ FDGYEDLLRK LEEMFDIKGE LCGSAKKWQV 540 VYTDNEDDMM MVGDDPWLEF CSVVRKIFIY TAEEVKKLSP KIGLPMNDEG KPSKLDSEAV 600 VNPEDQSSVV GPSC |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 4ldv_A | 0.0 | 1 | 302 | 56 | 357 | Auxin response factor 1 |
| 4ldw_A | 0.0 | 1 | 302 | 56 | 357 | Auxin response factor 1 |
| 4ldw_B | 0.0 | 1 | 302 | 56 | 357 | Auxin response factor 1 |
| Search in ModeBase | ||||||
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Lja.6764 | 0.0 | pod | ||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | DEVELOPMENTAL STAGE: Expressed in the sepals and carpels of young flower buds. At stage 10 of flower development, expression in the carpels becomes restricted to the style. Also expressed in anthers and filaments. At stage 13, expressed in the region at the top of the pedicel, including the abscission zone. {ECO:0000269|PubMed:16176952}. | |||||
| Uniprot | TISSUE SPECIFICITY: Expressed in the whole plant. {ECO:0000269|PubMed:10476078}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Auxin response factors (ARFs) are transcriptional factors that bind specifically to the DNA sequence 5'-TGTCTC-3' found in the auxin-responsive promoter elements (AuxREs). Seems to act as transcriptional repressor. Formation of heterodimers with Aux/IAA proteins may alter their ability to modulate early auxin response genes expression. Promotes flowering, stamen development, floral organ abscission and fruit dehiscence. Acts as repressor of IAA2, IAA3 and IAA7. {ECO:0000269|PubMed:12036261, ECO:0000269|PubMed:16176952}. | |||||
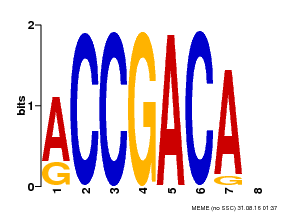
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00097 | PBM | Transfer from AT1G59750 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Lj0g3v0140239.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_003592874.1 | 0.0 | auxin response factor 1 | ||||
| Refseq | XP_024631839.1 | 0.0 | auxin response factor 1 | ||||
| Swissprot | Q8L7G0 | 0.0 | ARFA_ARATH; Auxin response factor 1 | ||||
| TrEMBL | G7IL56 | 0.0 | G7IL56_MEDTR; Auxin response factor | ||||
| STRING | AES63125 | 0.0 | (Medicago truncatula) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF6802 | 26 | 38 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G59750.4 | 0.0 | auxin response factor 1 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



