 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Lj0g3v0157669.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Loteae; Lotus
|
||||||||
| Family | ERF | ||||||||
| Protein Properties | Length: 276aa MW: 31050.8 Da PI: 5.6963 | ||||||||
| Description | ERF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 57.9 | 2.4e-18 | 31 | 80 | 2 | 55 |
AP2 2 gykGVrwdkkrgrWvAeIrdpsengkrkrfslgkfgtaeeAakaaiaarkkleg 55
y+GVr++ +g+WvAeIr+p++ r+r +lg+f taeeAa a+++a+ +l+g
Lj0g3v0157669.1 31 EYRGVRQRT-WGKWVAEIREPKK---RTRLWLGSFTTAEEAAMAYDEAAWRLYG 80
69****999.**********965...5*********************988877 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| CDD | cd00018 | 1.55E-31 | 30 | 90 | No hit | No description |
| PROSITE profile | PS51032 | 23.037 | 31 | 88 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 2.1E-37 | 31 | 94 | IPR001471 | AP2/ERF domain |
| Pfam | PF00847 | 5.5E-12 | 31 | 80 | IPR001471 | AP2/ERF domain |
| SuperFamily | SSF54171 | 1.83E-21 | 31 | 89 | IPR016177 | DNA-binding domain |
| Gene3D | G3DSA:3.30.730.10 | 3.4E-30 | 31 | 89 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 2.0E-10 | 32 | 43 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 2.0E-10 | 54 | 70 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 276 aa Download sequence Send to blast |
MDHTCKRSSA LKPWKKGPTR GKGGPQNASC EYRGVRQRTW GKWVAEIREP KKRTRLWLGS 60 FTTAEEAAMA YDEAAWRLYG PDAYLNLPHL QPSSNAAPIK SGKFKWLPSK NFMSMFPTCG 120 ALNVNAQPSV HFIHQRLQEL KQNAVVTQPP SKEEIMTVGS KNNAEKPSAE KDAQTLPEAM 180 LGDLQEKPQI DLHEFLQQLG ILKEERHSER ADSSGSSTVN EAVSRDDNDQ LGVFSDNSVN 240 WEALIAGIQE SEDIIQLEEA YDMNDELNFS TSIWNF |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1gcc_A | 4e-18 | 32 | 88 | 3 | 60 | ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional activator that binds specifically to the DNA sequence 5'-[AG]CCGAC-3'. Binding to the C-repeat/DRE element mediates high salinity-inducible transcription. {ECO:0000269|PubMed:11798174}. | |||||
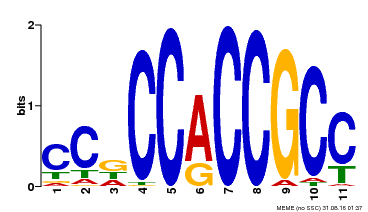
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00411 | DAP | Transfer from AT3G57600 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Lj0g3v0157669.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By high-salt stress. {ECO:0000269|PubMed:11798174}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_004493523.1 | 1e-146 | dehydration-responsive element-binding protein 2F | ||||
| Swissprot | Q9SVX5 | 3e-85 | DRE2F_ARATH; Dehydration-responsive element-binding protein 2F | ||||
| TrEMBL | A0A1S2XRS0 | 1e-145 | A0A1S2XRS0_CICAR; dehydration-responsive element-binding protein 2F | ||||
| STRING | XP_004493523.1 | 1e-146 | (Cicer arietinum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF5477 | 31 | 50 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G57600.1 | 3e-77 | ERF family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



