 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Lj0g3v0221649.2 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Loteae; Lotus
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 149aa MW: 16774.1 Da PI: 4.8067 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 37.8 | 4.3e-12 | 12 | 56 | 3 | 45 |
SS-HHHHHHHHHHHHHTTTT...-HHHHHHHHTTTS-HHHHHHHHH CS
Myb_DNA-binding 3 rWTteEdellvdavkqlGgg...tWktIartmgkgRtlkqcksrwq 45
W+ e d+ + +a + ++ + +W++Ia+ ++ g+tl+++k+++
Lj0g3v0221649.2 12 EWSREQDKAFENALATYPEDdsdRWEKIAADVP-GKTLEEIKHHYE 56
7********************************.**********96 PP
| |||||||
| 2 | Myb_DNA-binding | 26.2 | 1.8e-08 | 123 | 149 | 3 | 29 |
SS-HHHHHHHHHHHHHTTTT-HHHHHH CS
Myb_DNA-binding 3 rWTteEdellvdavkqlGggtWktIar 29
+WT+eE+ l++ + ++G+g+W++I+r
Lj0g3v0221649.2 123 AWTEEEHRLFLLGLDKYGKGDWRSISR 149
6************************97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51293 | 9.704 | 8 | 63 | IPR017884 | SANT domain |
| SMART | SM00717 | 1.8E-10 | 9 | 61 | IPR001005 | SANT/Myb domain |
| SuperFamily | SSF46689 | 4.59E-12 | 11 | 67 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 3.41E-9 | 12 | 59 | No hit | No description |
| Pfam | PF00249 | 1.7E-9 | 12 | 56 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 5.6E-6 | 12 | 56 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 12.693 | 115 | 149 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 5.17E-6 | 118 | 148 | IPR009057 | Homeodomain-like |
| Pfam | PF00249 | 3.8E-6 | 123 | 148 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 5.26E-6 | 123 | 149 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 1.1E-6 | 123 | 149 | IPR009057 | Homeodomain-like |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 149 aa Download sequence Send to blast |
MTVDEVGSSS SEWSREQDKA FENALATYPE DDSDRWEKIA ADVPGKTLEE IKHHYELLVD 60 DVNQIDSGCV PVPSYNSFSE GSRSHASDEG AGKKGGHPWN YNSESNHGTK ASRSDQERRK 120 GIAWTEEEHR LFLLGLDKYG KGDWRSISR |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 2cjj_A | 4e-19 | 9 | 77 | 7 | 75 | RADIALIS |
| Search in ModeBase | ||||||
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Lja.1022 | 0.0 | pod| root | ||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | TISSUE SPECIFICITY: Expressed in young seedlings, developing leaves, sepals and trichomes. {ECO:0000269|PubMed:26243618}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator that coordinates abscisic acid (ABA) biosynthesis and signaling-related genes via binding to the specific promoter motif 5'-(A/T)AACCAT-3'. Represses ABA-mediated salt (e.g. NaCl and KCl) stress tolerance. Regulates leaf shape and promotes vegetative growth. {ECO:0000269|PubMed:26243618}. | |||||
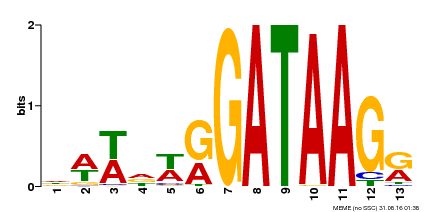
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00500 | DAP | Transfer from AT5G08520 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Lj0g3v0221649.2 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Induced by salicylic acid (SA) and gibberellic acid (GA) (PubMed:16463103). Triggered by dehydration and salt stress (PubMed:26243618). {ECO:0000269|PubMed:16463103, ECO:0000269|PubMed:26243618}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | - | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | BT146241 | 0.0 | BT146241.1 Lotus japonicus clone JCVI-FLLj-3K12 unknown mRNA. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_003532025.1 | 4e-92 | transcription factor SRM1 | ||||
| Refseq | XP_006585964.1 | 4e-92 | transcription factor SRM1 | ||||
| Refseq | XP_025985536.1 | 4e-92 | transcription factor SRM1 | ||||
| Refseq | XP_025985537.1 | 4e-92 | transcription factor SRM1 | ||||
| Refseq | XP_028245644.1 | 4e-92 | transcription factor SRM1-like | ||||
| Refseq | XP_028245645.1 | 4e-92 | transcription factor SRM1-like | ||||
| Refseq | XP_028245646.1 | 4e-92 | transcription factor SRM1-like | ||||
| Swissprot | Q9FNN6 | 5e-61 | SRM1_ARATH; Transcription factor SRM1 | ||||
| TrEMBL | A0A445JLL2 | 9e-91 | A0A445JLL2_GLYSO; Transcription factor SRM1 isoform A | ||||
| TrEMBL | K7L9L3 | 9e-91 | K7L9L3_SOYBN; Uncharacterized protein | ||||
| STRING | GLYMA08G40020.5 | 1e-91 | (Glycine max) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G08520.1 | 2e-63 | MYB family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



