 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Lj0g3v0342589.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Loteae; Lotus
|
||||||||
| Family | ARF | ||||||||
| Protein Properties | Length: 927aa MW: 102588 Da PI: 5.708 | ||||||||
| Description | ARF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | B3 | 73.9 | 2e-23 | 153 | 254 | 1 | 99 |
EEEE-..-HHHHTT-EE--HHH.HTT......---..--SEEEEEETTS-EEEEEE..EEETTEEEE-TTHHHHHHHHT--TT-EEEEEE-SSS CS
B3 1 ffkvltpsdvlksgrlvlpkkfaeeh......ggkkeesktltledesgrsWevkliyrkksgryvltkGWkeFvkangLkegDfvvFkldgrs 88
f+k+lt sd++++g +++p++ ae+ +++++ ++l+++d++ ++W++++iyr++++r++lt+GW+ Fv +++L++gD+v+F +++
Lj0g3v0342589.1 153 FCKTLTASDTSTHGGFSVPRRAAEKLfpqldyTIQPPT-QELVVRDLHDNTWTFRHIYRGQPKRHLLTTGWSLFVGSKRLRAGDSVLFI--RDE 243
99*********************999****98555555.59************************************************..456 PP
EE..EEEEE-S CS
B3 89 efelvvkvfrk 99
+l+v+v+r+
Lj0g3v0342589.1 244 ISQLRVGVRRA 254
7777****997 PP
| |||||||
| 2 | Auxin_resp | 115.7 | 4.2e-38 | 279 | 362 | 1 | 83 |
Auxin_resp 1 aahaastksvFevvYnPrastseFvvkvekvekalk.vkvsvGmRfkmafetedsserrlsGtvvgvsdldpvrWpnSkWrsLk 83
aahaa+++s+F+++YnPra++seFv++++k++ka++ +++svGmRf m+fete+s +rr++Gt+vg++d+dp+rWp+SkWr+++
Lj0g3v0342589.1 279 AAHAAANRSPFTIFYNPRACPSEFVIPLAKYRKAVYaSQLSVGMRFGMMFETEESGKRRYMGTIVGINDVDPLRWPGSKWRNIQ 362
79*********************************989*******************************************986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF101936 | 6.8E-46 | 142 | 282 | IPR015300 | DNA-binding pseudobarrel domain |
| Gene3D | G3DSA:2.40.330.10 | 2.9E-40 | 145 | 267 | IPR015300 | DNA-binding pseudobarrel domain |
| CDD | cd10017 | 5.23E-22 | 152 | 253 | No hit | No description |
| Pfam | PF02362 | 8.1E-21 | 153 | 254 | IPR003340 | B3 DNA binding domain |
| SMART | SM01019 | 4.1E-23 | 153 | 255 | IPR003340 | B3 DNA binding domain |
| PROSITE profile | PS50863 | 12.788 | 153 | 255 | IPR003340 | B3 DNA binding domain |
| Pfam | PF06507 | 2.4E-33 | 279 | 362 | IPR010525 | Auxin response factor |
| Pfam | PF02309 | 4.6E-11 | 818 | 908 | IPR033389 | AUX/IAA domain |
| PROSITE profile | PS51745 | 23.746 | 821 | 905 | IPR000270 | PB1 domain |
| SuperFamily | SSF54277 | 5.79E-8 | 834 | 900 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009733 | Biological Process | response to auxin | ||||
| GO:0009908 | Biological Process | flower development | ||||
| GO:0009942 | Biological Process | longitudinal axis specification | ||||
| GO:0010305 | Biological Process | leaf vascular tissue pattern formation | ||||
| GO:0048364 | Biological Process | root development | ||||
| GO:0048507 | Biological Process | meristem development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0016020 | Cellular Component | membrane | ||||
| GO:0042802 | Molecular Function | identical protein binding | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 927 aa Download sequence Send to blast |
MASVEEKIKT GGVGVGVGVG VVGGQTQTQN LVAEMKLLKD CGVRRTLNSE LWHACAGPLV 60 SLPQVGSLVY YFPQGHSEQV AASTRRTATS QIPNYPSLQS QLLCQVQNVT LHADKETDEI 120 YAQMSLQPVN SEKEVFPISD FGTKQSKHPS EFFCKTLTAS DTSTHGGFSV PRRAAEKLFP 180 QLDYTIQPPT QELVVRDLHD NTWTFRHIYR GQPKRHLLTT GWSLFVGSKR LRAGDSVLFI 240 RDEISQLRVG VRRANRQQTT LPSSVLSADS MHIGVLAAAA HAAANRSPFT IFYNPRACPS 300 EFVIPLAKYR KAVYASQLSV GMRFGMMFET EESGKRRYMG TIVGINDVDP LRWPGSKWRN 360 IQVEWDEPGC GDKQNRVSVW DIETPESLFI FPSLTSSLKR PLQSGLLENE WGTLLRKPFI 420 KAPESGAMEL SSSIPNLYQE QLMKMLFKPQ AVNNNGAFPS LMQQESAATR GPLQVDNQNV 480 HLASTGSMTL KNPCSQSSQN SQTTLLKSDQ PEKLHPQPRI DNHLTSGTVT DNKSRLESEV 540 LDHVLDFPSM EGCKIEKMVT NPINPQNLAS HLTINNQNQN PLLPQSSTWP MQHTQLESSL 600 SYPQVIDMSQ SDSAIVNGTL PQLDIEEWMA YSSCQPFAGQ NRSNGPFSDF QEHASLQAQV 660 VNNPSLTSTN QEVLDHYVKN LKFFSQADQL TSIYGFNGIS SSNNLRDLSS ESNNQSEICV 720 NVDVSNSVNT TTVDPSTTSS TILDEFCTMK ERDFQHPQEC MVGNLSCSQD GQSQITSASL 780 AESHAFCLRD NSGGTSSSHV DFDDSSLLQN NSWQQGSTPI RTYTKVQKAG SVGRSIDVTT 840 FKNYEELIRA IECMFGLDGL LNDTKGSGWK LVYVDYESDV LLVGDDPWEE FVGCVRCIRI 900 LSPSEVQQMS EEGMKLLNSG ALQGINV |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 4ldu_A | 0.0 | 1 | 387 | 5 | 392 | Auxin response factor 5 |
| Search in ModeBase | ||||||
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Lja.20685 | 0.0 | pod | ||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | DEVELOPMENTAL STAGE: In early embryo and during organ development. | |||||
| Uniprot | TISSUE SPECIFICITY: Expressed in the whole plant with a lower expression in leaves. Detected in embryo axis, provascular tissues, procambium and some differentiated vascular regions of mature organs. {ECO:0000269|PubMed:10476078}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Auxin response factors (ARFs) are transcriptional factors that bind specifically to the DNA sequence 5'-TGTCTC-3' found in the auxin-responsive promoter elements (AuxREs). Seems to act as transcriptional activator. Formation of heterodimers with Aux/IAA proteins may alter their ability to modulate early auxin response genes expression. Mediates embryo axis formation and vascular tissues differentiation. Functionally redundant with ARF7. May be necessary to counteract AMP1 activity. {ECO:0000269|PubMed:12036261, ECO:0000269|PubMed:14973283, ECO:0000269|PubMed:17553903}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00153 | DAP | Transfer from AT1G19850 | Download |
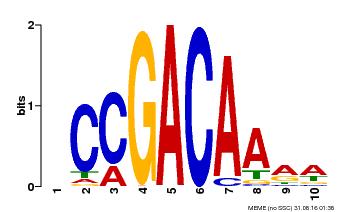
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Lj0g3v0342589.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020233090.1 | 0.0 | auxin response factor 5 | ||||
| Swissprot | P93024 | 0.0 | ARFE_ARATH; Auxin response factor 5 | ||||
| TrEMBL | A0A445H8Z3 | 0.0 | A0A445H8Z3_GLYSO; Auxin response factor | ||||
| STRING | GLYMA14G40540.1 | 0.0 | (Glycine max) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G19850.1 | 0.0 | ARF family protein | ||||



