 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Lj0g3v0342589.2 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Loteae; Lotus
|
||||||||
| Family | ARF | ||||||||
| Protein Properties | Length: 958aa MW: 106317 Da PI: 5.6162 | ||||||||
| Description | ARF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | B3 | 73.8 | 2e-23 | 184 | 285 | 1 | 99 |
EEEE-..-HHHHTT-EE--HHH.HTT......---..--SEEEEEETTS-EEEEEE..EEETTEEEE-TTHHHHHHHHT--TT-EEEEEE-SSS CS
B3 1 ffkvltpsdvlksgrlvlpkkfaeeh......ggkkeesktltledesgrsWevkliyrkksgryvltkGWkeFvkangLkegDfvvFkldgrs 88
f+k+lt sd++++g +++p++ ae+ +++++ ++l+++d++ ++W++++iyr++++r++lt+GW+ Fv +++L++gD+v+F +++
Lj0g3v0342589.2 184 FCKTLTASDTSTHGGFSVPRRAAEKLfpqldyTIQPPT-QELVVRDLHDNTWTFRHIYRGQPKRHLLTTGWSLFVGSKRLRAGDSVLFI--RDE 274
99*********************999****98555555.59************************************************..456 PP
EE..EEEEE-S CS
B3 89 efelvvkvfrk 99
+l+v+v+r+
Lj0g3v0342589.2 275 ISQLRVGVRRA 285
7777****997 PP
| |||||||
| 2 | Auxin_resp | 115.6 | 4.4e-38 | 310 | 393 | 1 | 83 |
Auxin_resp 1 aahaastksvFevvYnPrastseFvvkvekvekalk.vkvsvGmRfkmafetedsserrlsGtvvgvsdldpvrWpnSkWrsLk 83
aahaa+++s+F+++YnPra++seFv++++k++ka++ +++svGmRf m+fete+s +rr++Gt+vg++d+dp+rWp+SkWr+++
Lj0g3v0342589.2 310 AAHAAANRSPFTIFYNPRACPSEFVIPLAKYRKAVYaSQLSVGMRFGMMFETEESGKRRYMGTIVGINDVDPLRWPGSKWRNIQ 393
79*********************************989*******************************************986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF101936 | 7.06E-46 | 173 | 313 | IPR015300 | DNA-binding pseudobarrel domain |
| Gene3D | G3DSA:2.40.330.10 | 3.1E-40 | 176 | 298 | IPR015300 | DNA-binding pseudobarrel domain |
| CDD | cd10017 | 5.54E-22 | 183 | 284 | No hit | No description |
| Pfam | PF02362 | 8.5E-21 | 184 | 285 | IPR003340 | B3 DNA binding domain |
| PROSITE profile | PS50863 | 12.788 | 184 | 286 | IPR003340 | B3 DNA binding domain |
| SMART | SM01019 | 4.1E-23 | 184 | 286 | IPR003340 | B3 DNA binding domain |
| Pfam | PF06507 | 2.5E-33 | 310 | 393 | IPR010525 | Auxin response factor |
| Pfam | PF02309 | 4.8E-11 | 849 | 939 | IPR033389 | AUX/IAA domain |
| PROSITE profile | PS51745 | 23.746 | 852 | 936 | IPR000270 | PB1 domain |
| SuperFamily | SSF54277 | 5.98E-8 | 865 | 931 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009733 | Biological Process | response to auxin | ||||
| GO:0009908 | Biological Process | flower development | ||||
| GO:0009942 | Biological Process | longitudinal axis specification | ||||
| GO:0010305 | Biological Process | leaf vascular tissue pattern formation | ||||
| GO:0048364 | Biological Process | root development | ||||
| GO:0048507 | Biological Process | meristem development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0016020 | Cellular Component | membrane | ||||
| GO:0042802 | Molecular Function | identical protein binding | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 958 aa Download sequence Send to blast |
MASVEEKIKT GGVGVGVGVG VVGGQTQTQN LVAEMKLLKD CGLFPFFVCI EETKINLLLW 60 FFTCLFMLLI FSGVRRTLNS ELWHACAGPL VSLPQVGSLV YYFPQGHSEQ VAASTRRTAT 120 SQIPNYPSLQ SQLLCQVQNV TLHADKETDE IYAQMSLQPV NSEKEVFPIS DFGTKQSKHP 180 SEFFCKTLTA SDTSTHGGFS VPRRAAEKLF PQLDYTIQPP TQELVVRDLH DNTWTFRHIY 240 RGQPKRHLLT TGWSLFVGSK RLRAGDSVLF IRDEISQLRV GVRRANRQQT TLPSSVLSAD 300 SMHIGVLAAA AHAAANRSPF TIFYNPRACP SEFVIPLAKY RKAVYASQLS VGMRFGMMFE 360 TEESGKRRYM GTIVGINDVD PLRWPGSKWR NIQVEWDEPG CGDKQNRVSV WDIETPESLF 420 IFPSLTSSLK RPLQSGLLEN EWGTLLRKPF IKAPESGAME LSSSIPNLYQ EQLMKMLFKP 480 QAVNNNGAFP SLMQQESAAT RGPLQVDNQN VHLASTGSMT LKNPCSQSSQ NSQTTLLKSD 540 QPEKLHPQPR IDNHLTSGTV TDNKSRLESE VLDHVLDFPS MEGCKIEKMV TNPINPQNLA 600 SHLTINNQNQ NPLLPQSSTW PMQHTQLESS LSYPQVIDMS QSDSAIVNGT LPQLDIEEWM 660 AYSSCQPFAG QNRSNGPFSD FQEHASLQAQ VVNNPSLTST NQEVLDHYVK NLKFFSQADQ 720 LTSIYGFNGI SSSNNLRDLS SESNNQSEIC VNVDVSNSVN TTTVDPSTTS STILDEFCTM 780 KERDFQHPQE CMVGNLSCSQ DGQSQITSAS LAESHAFCLR DNSGGTSSSH VDFDDSSLLQ 840 NNSWQQGSTP IRTYTKVQKA GSVGRSIDVT TFKNYEELIR AIECMFGLDG LLNDTKGSGW 900 KLVYVDYESD VLLVGDDPWE EFVGCVRCIR ILSPSEVQQM SEEGMKLLNS GALQGINV |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 4ldu_A | 0.0 | 1 | 418 | 5 | 392 | Auxin response factor 5 |
| Search in ModeBase | ||||||
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Lja.20685 | 0.0 | pod | ||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | DEVELOPMENTAL STAGE: In early embryo and during organ development. | |||||
| Uniprot | TISSUE SPECIFICITY: Expressed in the whole plant with a lower expression in leaves. Detected in embryo axis, provascular tissues, procambium and some differentiated vascular regions of mature organs. {ECO:0000269|PubMed:10476078}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Auxin response factors (ARFs) are transcriptional factors that bind specifically to the DNA sequence 5'-TGTCTC-3' found in the auxin-responsive promoter elements (AuxREs). Seems to act as transcriptional activator. Formation of heterodimers with Aux/IAA proteins may alter their ability to modulate early auxin response genes expression. Mediates embryo axis formation and vascular tissues differentiation. Functionally redundant with ARF7. May be necessary to counteract AMP1 activity. {ECO:0000269|PubMed:12036261, ECO:0000269|PubMed:14973283, ECO:0000269|PubMed:17553903}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00153 | DAP | Transfer from AT1G19850 | Download |
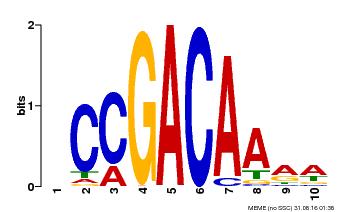
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Lj0g3v0342589.2 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_027331369.1 | 0.0 | auxin response factor 5-like isoform X1 | ||||
| Swissprot | P93024 | 0.0 | ARFE_ARATH; Auxin response factor 5 | ||||
| TrEMBL | A0A445H8Z3 | 0.0 | A0A445H8Z3_GLYSO; Auxin response factor | ||||
| STRING | GLYMA14G40540.1 | 0.0 | (Glycine max) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF8991 | 32 | 41 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G19850.1 | 0.0 | ARF family protein | ||||



