 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Lj1g3v0175780.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Loteae; Lotus
|
||||||||
| Family | MIKC_MADS | ||||||||
| Protein Properties | Length: 243aa MW: 27660.4 Da PI: 9.5344 | ||||||||
| Description | MIKC_MADS family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SRF-TF | 87.5 | 7.5e-28 | 10 | 59 | 2 | 51 |
---SHHHHHHHHHHHHHHHHHHHHHHHHHHT-EEEEEEE-TTSEEEEEE- CS
SRF-TF 2 rienksnrqvtfskRrngilKKAeELSvLCdaevaviifsstgklyeyss 51
ri+n++ rqvtfskRrng+lKKA+EL +LCdaev v+ifsstg+lye++s
Lj1g3v0175780.1 10 RIDNSTSRQVTFSKRRNGLLKKAKELAILCDAEVGVVIFSSTGRLYEFAS 59
8***********************************************86 PP
| |||||||
| 2 | K-box | 80.2 | 5e-27 | 78 | 170 | 8 | 98 |
K-box 8 s..leeakaeslqqelakLkkeienLqreqRhllGedLesLslkeLqqLeqqLekslkkiRskKnellleqieelqkkekelqeenkaLrkkl 98
+ ++++ + +q+e+a L+++++ Lq+++R+++Ge+L+ L++keLq+Le+qLe sl +R kK++ll+++i+el +k + +++en +L kk+
Lj1g3v0175780.1 78 QlgSSTSEIKLWQREAAMLRQQLHCLQESHRQIMGEELSGLTVKELQSLESQLEISLHGVRMKKDQLLMDEIQELNRKGNLIHQENVELYKKV 170
2347889999********************************************************************************997 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50066 | 31.763 | 1 | 61 | IPR002100 | Transcription factor, MADS-box |
| SMART | SM00432 | 3.0E-39 | 1 | 60 | IPR002100 | Transcription factor, MADS-box |
| SuperFamily | SSF55455 | 4.71E-32 | 2 | 85 | IPR002100 | Transcription factor, MADS-box |
| CDD | cd00265 | 1.72E-43 | 2 | 76 | No hit | No description |
| PROSITE pattern | PS00350 | 0 | 3 | 57 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 3.2E-28 | 3 | 23 | IPR002100 | Transcription factor, MADS-box |
| Pfam | PF00319 | 1.4E-25 | 10 | 57 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 3.2E-28 | 23 | 38 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 3.2E-28 | 38 | 59 | IPR002100 | Transcription factor, MADS-box |
| Pfam | PF01486 | 2.6E-26 | 83 | 170 | IPR002487 | Transcription factor, K-box |
| PROSITE profile | PS51297 | 15.91 | 86 | 176 | IPR002487 | Transcription factor, K-box |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0010440 | Biological Process | stomatal lineage progression | ||||
| GO:0048574 | Biological Process | long-day photoperiodism, flowering | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0008134 | Molecular Function | transcription factor binding | ||||
| GO:0042803 | Molecular Function | protein homodimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 243 aa Download sequence Send to blast |
MGRGKIVIRR IDNSTSRQVT FSKRRNGLLK KAKELAILCD AEVGVVIFSS TGRLYEFASS 60 SMKSVIDRYN KSKEEQNQLG SSTSEIKLWQ REAAMLRQQL HCLQESHRQI MGEELSGLTV 120 KELQSLESQL EISLHGVRMK KDQLLMDEIQ ELNRKGNLIH QENVELYKKV NLISQENMEL 180 KEKGYRTKDW NVPNSNSVLT SGQSTGEDLQ VPVNLQLSQP QPQQYLSPSG TTKLGYDLSS 240 FSR |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5f28_A | 6e-20 | 1 | 74 | 1 | 74 | MEF2C |
| 5f28_B | 6e-20 | 1 | 74 | 1 | 74 | MEF2C |
| 5f28_C | 6e-20 | 1 | 74 | 1 | 74 | MEF2C |
| 5f28_D | 6e-20 | 1 | 74 | 1 | 74 | MEF2C |
| Search in ModeBase | ||||||
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Lja.23265 | 0.0 | root | ||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | TISSUE SPECIFICITY: Ubiquitous. {ECO:0000269|PubMed:14701936}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00410 | DAP | Transfer from AT3G57230 | Download |
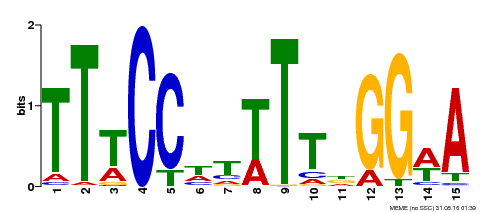
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Lj1g3v0175780.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | BT144757 | 0.0 | BT144757.1 Lotus japonicus clone JCVI-FLLj-15G19 unknown mRNA. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_027331266.1 | 1e-146 | MADS-box transcription factor 23-like | ||||
| Swissprot | Q6EP49 | 1e-104 | MAD27_ORYSJ; MADS-box transcription factor 27 | ||||
| TrEMBL | I3SWA9 | 1e-145 | I3SWA9_LOTJA; Uncharacterized protein | ||||
| STRING | GLYMA14G36221.2 | 1e-141 | (Glycine max) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF891 | 25 | 60 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G57230.1 | 8e-93 | AGAMOUS-like 16 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



