 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Lj1g3v0513130.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Loteae; Lotus
|
||||||||
| Family | Dof | ||||||||
| Protein Properties | Length: 525aa MW: 58187.4 Da PI: 6.4522 | ||||||||
| Description | Dof family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-Dof | 122.4 | 1.5e-38 | 173 | 233 | 2 | 62 |
zf-Dof 2 kekalkcprCdstntkfCyynnyslsqPryfCkaCrryWtkGGalrnvPvGggrrknkkss 62
++k l+cprC+s++tkfCyynny+++qPryfCkaC+ryWt+GG++r vPvG+grrknk+ss
Lj1g3v0513130.1 173 PDKLLPCPRCKSMDTKFCYYNNYNVNQPRYFCKACQRYWTAGGTMRHVPVGAGRRKNKNSS 233
78999*****************************************************987 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF02701 | 8.8E-32 | 175 | 231 | IPR003851 | Zinc finger, Dof-type |
| PROSITE profile | PS50884 | 28.164 | 177 | 231 | IPR003851 | Zinc finger, Dof-type |
| PROSITE pattern | PS01361 | 0 | 179 | 215 | IPR003851 | Zinc finger, Dof-type |
| ProDom | PD007478 | 1.0E-20 | 184 | 230 | IPR003851 | Zinc finger, Dof-type |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 525 aa Download sequence Send to blast |
MLPLPTETKN PNKTETLKTR SLAVIFRISR NRLPRFPQPS PRFPRTLFGS RENAGNEGSC 60 HQAFRPEDSL YPSTAAAIDV DKEEEEEDDV DRSEPEIDKV DQGDKDPPAE KVTEKKKEAD 120 PPPNTVAETK DIQNPKTPPI DEETANSETG KSENDQNDEN NNINSDDKTL KKPDKLLPCP 180 RCKSMDTKFC YYNNYNVNQP RYFCKACQRY WTAGGTMRHV PVGAGRRKNK NSSSHYDHIT 240 VSEARQAARI GAANGTHHAL KSNGRVLSFG IDPPICDSVA SVLNLGDSKK ALSGTRNRFH 300 NHTSEEQKIP VSGENGEDCS SSITVSNSME ETGKSTIQET FLPNNGFVPQ QVQCFSSVPW 360 PYPWGSGVPS QPQPALCPPG FFYPAAPMWN WSVPWFSSHT SAPNQMSPSS GSNSPTLGKH 420 SRNDDDMVKE QDLQQQEEES SQRHRNGCVL VPKTLRIDDP DEAAKSSIWA TLGIKNECVN 480 VSGGGMFKAF QSKKKEEKNH VLETSSVLLA NPAALSRSLN FHENT |
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Lja.22108 | 0.0 | protoplast | ||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | TISSUE SPECIFICITY: Expressed in the vasculature of cotyledons and hypocotyls, leaves and roots. {ECO:0000269|PubMed:19619493}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
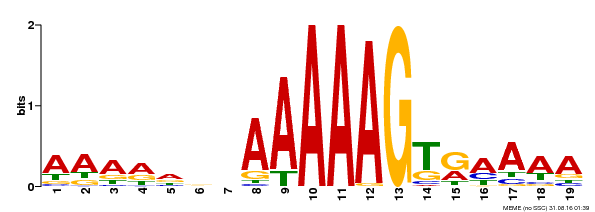
| UniProt | Transcription factor that binds specifically to a 5'-AA[AG]G-3' consensus core sequence (By similarity). Regulates a photoperiodic flowering response. Transcriptional repressor of 'CONSTANS' expression. {ECO:0000250, ECO:0000269|PubMed:19619493}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00394 | DAP | Transfer from AT3G47500 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Lj1g3v0513130.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Circadian-regulation. Highly expressed at the beginning of the light period, then decreases, reaching a minimum between 16 and 29 hours after dawn before rising again at the end of the day. {ECO:0000269|PubMed:19619493}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_027334746.1 | 0.0 | cyclic dof factor 3-like | ||||
| Swissprot | Q8LFV3 | 1e-110 | CDF3_ARATH; Cyclic dof factor 3 | ||||
| TrEMBL | A0A1S3TR26 | 1e-180 | A0A1S3TR26_VIGRR; cyclic dof factor 3 | ||||
| STRING | GLYMA06G20950.1 | 1e-177 | (Glycine max) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF625 | 34 | 147 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G47500.1 | 1e-95 | cycling DOF factor 3 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



