 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Lj2g3v1989210.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Loteae; Lotus
|
||||||||
| Family | ERF | ||||||||
| Protein Properties | Length: 295aa MW: 33229.5 Da PI: 6.8034 | ||||||||
| Description | ERF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 62.8 | 7.6e-20 | 137 | 187 | 2 | 55 |
AP2 2 gykGVrwdkkrgrWvAeIrdpsengkrkrfslgkfgtaeeAakaaiaarkkleg 55
+y+GVr+++ +g+++AeIrdp+++g r++lg+f+ta eAaka+++a+ ++g
Lj2g3v1989210.1 137 HYRGVRQRP-WGKFAAEIRDPNKRG--SRVWLGTFDTAIEAAKAYDRAAFTFRG 187
8********.**********96655..**********************99987 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:3.30.730.10 | 2.9E-32 | 136 | 196 | IPR001471 | AP2/ERF domain |
| CDD | cd00018 | 5.49E-32 | 136 | 195 | No hit | No description |
| SMART | SM00380 | 1.4E-37 | 137 | 201 | IPR001471 | AP2/ERF domain |
| Pfam | PF00847 | 7.6E-14 | 137 | 187 | IPR001471 | AP2/ERF domain |
| PROSITE profile | PS51032 | 24.144 | 137 | 195 | IPR001471 | AP2/ERF domain |
| SuperFamily | SSF54171 | 2.16E-22 | 137 | 196 | IPR016177 | DNA-binding domain |
| PRINTS | PR00367 | 1.8E-10 | 138 | 149 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 1.8E-10 | 161 | 177 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009409 | Biological Process | response to cold | ||||
| GO:0010200 | Biological Process | response to chitin | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 295 aa Download sequence Send to blast |
MANPDEVCAL YRIKLHLLGE LSPIPIRTTH THTHTHDDDF AFQFDLPNPN SSQSQSQSQS 60 FTSDPIFEFD CKPIIFDIET ESPKPIPVHK KQPALSRRKS LEISLPKKTT EWIQFGEVAP 120 EPSEASAKPE VEVEKKHYRG VRQRPWGKFA AEIRDPNKRG SRVWLGTFDT AIEAAKAYDR 180 AAFTFRGSKA VLNFPLEAGV VDRKHSREEE AEEAVKRQKT VESEIINRIR DTLLTPPSSS 240 GDVKGLFTVE SDINRIKETS LTPSSWTGFW DNFTVAPLSP LSPHPALGFP QLMVV |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 2gcc_A | 1e-29 | 136 | 196 | 4 | 64 | ATERF1 |
| 3gcc_A | 1e-29 | 136 | 196 | 4 | 64 | ATERF1 |
| Search in ModeBase | ||||||
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Lja.2990 | 7e-97 | cell culture| pod| protoplast| root | ||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00549 | DAP | Transfer from AT5G47230 | Download |
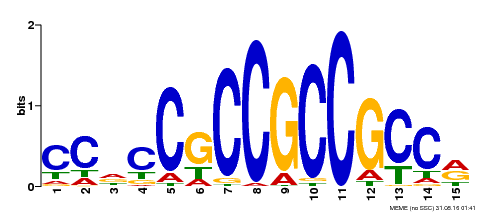
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Lj2g3v1989210.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | KT031253 | 2e-53 | KT031253.1 Glycine max clone HN_CCL_99 AP2-EREBP transcription factor (Glyma05g05130.1_like1) mRNA, partial cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | NP_001304366.2 | 3e-78 | ethylene-responsive transcription factor 5-like protein | ||||
| Refseq | XP_014507066.1 | 4e-78 | ethylene-responsive transcription factor 5 | ||||
| Refseq | XP_028186914.1 | 3e-78 | ethylene-responsive transcription factor 5-like | ||||
| TrEMBL | A0A1S3ULY1 | 1e-76 | A0A1S3ULY1_VIGRR; ethylene-responsive transcription factor 5 | ||||
| TrEMBL | A0A445HWI8 | 7e-77 | A0A445HWI8_GLYSO; Ethylene-responsive transcription factor 5 | ||||
| TrEMBL | C6TEG4 | 7e-77 | C6TEG4_SOYBN; Uncharacterized protein | ||||
| TrEMBL | I1LGT2 | 7e-77 | I1LGT2_SOYBN; Uncharacterized protein | ||||
| STRING | GLYMA11G03900.1 | 1e-77 | (Glycine max) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF1085 | 34 | 109 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G47230.1 | 2e-48 | ethylene responsive element binding factor 5 | ||||



