 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Lj4g3v1561700.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Loteae; Lotus
|
||||||||
| Family | E2F/DP | ||||||||
| Protein Properties | Length: 383aa MW: 42947.1 Da PI: 9.6185 | ||||||||
| Description | E2F/DP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | E2F_TDP | 75.9 | 4e-24 | 22 | 87 | 1 | 71 |
E2F_TDP 1 rkeksLrlltqkflkllekseegivtlnevakeLvsedvknkrRRiYDilNVLealnliekkekneirwkg 71
rk+ksL+ll+++fl+l+++ + + l+++a++L v +rRRiYDi+NVLe+++++++k+kn+++wkg
Lj4g3v1561700.1 22 RKQKSLGLLCTNFLSLYNRDNIHLIGLDDAASRL---GV--ERRRIYDIVNVLESIGVLSRKAKNQYTWKG 87
79*****************999************...**..****************************98 PP
| |||||||
| 2 | E2F_TDP | 75.5 | 5.2e-24 | 156 | 236 | 1 | 71 |
E2F_TDP 1 rkeksLrlltqkflkllekseegivtlnevakeLvse.....dvknkrRRiYDilNVLealnliek.....kekneirwkg 71
r+eksL lltq+f+kl+ +s+ ++++l+++ak L+ + +++k+RR+YDi+NVL+++ liek ++k+++rw g
Lj4g3v1561700.1 156 RREKSLALLTQNFVKLFLCSNVEMISLDDAAKLLLGDahntsIMRTKVRRLYDIANVLSSMSLIEKthtkdTRKPAFRWLG 236
689**************************************999***********************999999******87 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.10 | 5.1E-26 | 18 | 88 | IPR011991 | Winged helix-turn-helix DNA-binding domain |
| SuperFamily | SSF46785 | 2.33E-16 | 21 | 85 | IPR011991 | Winged helix-turn-helix DNA-binding domain |
| SMART | SM01372 | 1.2E-28 | 22 | 87 | IPR003316 | E2F/DP family, winged-helix DNA-binding domain |
| Pfam | PF02319 | 1.1E-20 | 23 | 87 | IPR003316 | E2F/DP family, winged-helix DNA-binding domain |
| Gene3D | G3DSA:1.10.10.10 | 6.6E-24 | 152 | 238 | IPR011991 | Winged helix-turn-helix DNA-binding domain |
| SMART | SM01372 | 4.6E-32 | 156 | 236 | IPR003316 | E2F/DP family, winged-helix DNA-binding domain |
| Pfam | PF02319 | 6.4E-19 | 158 | 236 | IPR003316 | E2F/DP family, winged-helix DNA-binding domain |
| SuperFamily | SSF46785 | 5.83E-13 | 158 | 237 | IPR011991 | Winged helix-turn-helix DNA-binding domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0032876 | Biological Process | negative regulation of DNA endoreduplication | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005667 | Cellular Component | transcription factor complex | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 383 aa Download sequence Send to blast |
MAAAAPPSDP AAASSKHHTY NRKQKSLGLL CTNFLSLYNR DNIHLIGLDD AASRLGVERR 60 RIYDIVNVLE SIGVLSRKAK NQYTWKGFRA IPLALQELKD EGLREKSSDF DGGNDSGKVS 120 DDEDDEEGFG NPSTGSQNEK SMSNSTLPRS MKNDNRREKS LALLTQNFVK LFLCSNVEMI 180 SLDDAAKLLL GDAHNTSIMR TKVRRLYDIA NVLSSMSLIE KTHTKDTRKP AFRWLGLKGR 240 TGNESVSTSN LNESRKRAFG TDVTNISFAR NRTDLFMNGD FGQNSKKQKI LENDGVLSQA 300 DKSNLQQDTK QTYQFGPFAP AYVSKAGNSE NSNVKQVHDW ESLATAHCPR YQNQALKELF 360 SHYMEAWRSW YSEVAGKRSI QTL |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 4yo2_A | 8e-42 | 22 | 228 | 4 | 228 | Transcription factor E2F8 |
| Search in ModeBase | ||||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | DEVELOPMENTAL STAGE: Expressed in a cell cycle-dependent manner. Not detected during early S phase. Expressed at both the G1/S and S/G2 transitions, with a peak during G2. {ECO:0000269|PubMed:11786543, ECO:0000269|PubMed:18787127}. | |||||
| Uniprot | TISSUE SPECIFICITY: Expressed exclusively in mitotically dividing cells. Highly expressed in young leaves and mature flowers. Lower expression in young stalk and in young and mature flowers. {ECO:0000269|PubMed:11867638, ECO:0000269|PubMed:15649366}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Inhibitor of E2F-dependent activation of gene expression. Binds specifically the E2 recognition site without interacting with DP proteins and prevents transcription activation by E2F/DP heterodimers. Controls the timing of endocycle onset and inhibits endoreduplication. {ECO:0000269|PubMed:11786543, ECO:0000269|PubMed:11867638, ECO:0000269|PubMed:15649366, ECO:0000269|PubMed:18787127}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00397 | DAP | Transfer from AT3G48160 | Download |
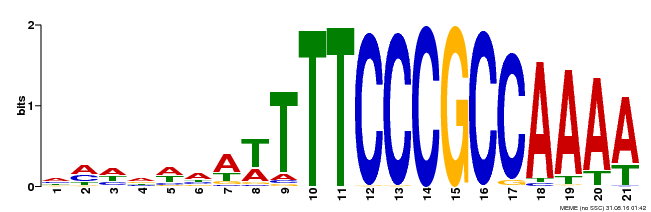
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Lj4g3v1561700.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AP009716 | 0.0 | AP009716.1 Lotus japonicus genomic DNA, chromosome 4, clone: LjT30K03, TM1793, complete sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_027368833.1 | 0.0 | E2F transcription factor-like E2FE | ||||
| Swissprot | Q8LSZ4 | 1e-127 | E2FE_ARATH; E2F transcription factor-like E2FE | ||||
| TrEMBL | A0A1S2YRL5 | 0.0 | A0A1S2YRL5_CICAR; E2F transcription factor-like E2FE isoform X1 | ||||
| STRING | XP_004508820.1 | 0.0 | (Cicer arietinum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF3668 | 33 | 66 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G48160.2 | 1e-119 | DP-E2F-like 1 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



