 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Lj4g3v2881950.2 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Loteae; Lotus
|
||||||||
| Family | ARF | ||||||||
| Protein Properties | Length: 487aa MW: 54322.1 Da PI: 7.9697 | ||||||||
| Description | ARF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | B3 | 76 | 4.3e-24 | 158 | 259 | 1 | 99 |
EEEE-..-HHHHTT-EE--HHH.HTT......---..--SEEEEEETTS-EEEEEE..EEETTEEEE-TTHHHHHHHHT--TT-EEEEEE-SSS CS
B3 1 ffkvltpsdvlksgrlvlpkkfaeeh......ggkkeesktltledesgrsWevkliyrkksgryvltkGWkeFvkangLkegDfvvFkldgrs 88
f+k+lt sd++++g +++ +++a+e+ + k++ +++l+ +d++g +W++++i+r++++r++l++GW+ Fv++++L +gD ++F + +
Lj4g3v2881950.2 158 FCKTLTASDTSTHGGFSVLRRHADEClppldmS-KQPPTQELVAKDLHGSEWRFRHIFRGQPRRHLLQSGWSVFVSSKRLVAGDAFIFL--RGE 248
99*****************************63.4445569************************************************..449 PP
EE..EEEEE-S CS
B3 89 efelvvkvfrk 99
++el+v+v+r+
Lj4g3v2881950.2 249 NGELRVGVRRA 259
999*****996 PP
| |||||||
| 2 | Auxin_resp | 116.2 | 2.8e-38 | 284 | 366 | 1 | 83 |
Auxin_resp 1 aahaastksvFevvYnPrastseFvvkvekvekalkvkvsvGmRfkmafetedsserrlsGtvvgvsdldpvrWpnSkWrsLk 83
a+ha t+++F+v+Y+Pr+s++eF+v++++++++lk+++++GmRfkm+fe+e+++e+r++Gt+vg++d+d+ +Wp+SkWr+Lk
Lj4g3v2881950.2 284 AWHANLTGTMFTVYYKPRTSPAEFIVPYDQYMESLKNNYTIGMRFKMRFEGEEAPEQRFTGTIVGIEDADSKSWPKSKWRCLK 366
799999***************************************************************************96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF101936 | 2.49E-39 | 154 | 287 | IPR015300 | DNA-binding pseudobarrel domain |
| Gene3D | G3DSA:2.40.330.10 | 9.0E-42 | 155 | 273 | IPR015300 | DNA-binding pseudobarrel domain |
| CDD | cd10017 | 5.95E-22 | 156 | 258 | No hit | No description |
| SMART | SM01019 | 5.8E-23 | 158 | 260 | IPR003340 | B3 DNA binding domain |
| PROSITE profile | PS50863 | 11.956 | 158 | 260 | IPR003340 | B3 DNA binding domain |
| Pfam | PF02362 | 5.0E-22 | 158 | 259 | IPR003340 | B3 DNA binding domain |
| Pfam | PF06507 | 6.0E-36 | 284 | 366 | IPR010525 | Auxin response factor |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0008285 | Biological Process | negative regulation of cell proliferation | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0009911 | Biological Process | positive regulation of flower development | ||||
| GO:0010047 | Biological Process | fruit dehiscence | ||||
| GO:0010150 | Biological Process | leaf senescence | ||||
| GO:0010227 | Biological Process | floral organ abscission | ||||
| GO:0045892 | Biological Process | negative regulation of transcription, DNA-templated | ||||
| GO:0048481 | Biological Process | plant ovule development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 487 aa Download sequence Send to blast |
MATSEVTNKG NSVNGMGDTN YNNNNNDGSR NGAGEGQSGS SSSPTRDAET ALYRELWHAC 60 AGPLVTVPRE GERVFYFPQG HIEQVEASTN QVAELHMPVY DLPPKILCRV INVMLKAEPD 120 TDEVFAQVTL LPEPNQDENA VEKEARLPPP PRFHVHSFCK TLTASDTSTH GGFSVLRRHA 180 DECLPPLDMS KQPPTQELVA KDLHGSEWRF RHIFRGQPRR HLLQSGWSVF VSSKRLVAGD 240 AFIFLRGENG ELRVGVRRAM RQQGNVSSSV ISSHSMHLGV LATAWHANLT GTMFTVYYKP 300 RTSPAEFIVP YDQYMESLKN NYTIGMRFKM RFEGEEAPEQ RFTGTIVGIE DADSKSWPKS 360 KWRCLKVRWD ETSNIPRPER VSPWKIEPAL APPAMNPLIM PRPKRPRSNV VPSSPDSSIL 420 TREASSKVNA DPLPASGFPR VLQGQESSTL RGNFAESIES NTAEKSVPWP RAADDEKIDA 480 ASTSRRY |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 4ldv_A | 0.0 | 40 | 388 | 7 | 354 | Auxin response factor 1 |
| 4ldw_A | 0.0 | 40 | 388 | 7 | 354 | Auxin response factor 1 |
| 4ldw_B | 0.0 | 40 | 388 | 7 | 354 | Auxin response factor 1 |
| 4ldx_A | 0.0 | 40 | 388 | 7 | 354 | Auxin response factor 1 |
| 4ldx_B | 0.0 | 40 | 388 | 7 | 354 | Auxin response factor 1 |
| Search in ModeBase | ||||||
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Lja.1056 | 0.0 | pod | ||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | DEVELOPMENTAL STAGE: Expressed in the sepals and carpels of young flower buds. At stage 10 of flower development, expression in the carpels becomes restricted to the style. Also expressed in anthers and filaments. At stage 13, expressed in the region at the top of the pedicel, including the abscission zone. Expressed in developing siliques. {ECO:0000269|PubMed:15960614, ECO:0000269|PubMed:16176952}. | |||||
| Uniprot | TISSUE SPECIFICITY: Expressed in the whole plant. {ECO:0000269|PubMed:10476078, ECO:0000269|PubMed:15960614}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Auxin response factors (ARFs) are transcriptional factors that bind specifically to the DNA sequence 5'-TGTCTC-3' found in the auxin-responsive promoter elements (AuxREs). Could act as transcriptional activator or repressor. Formation of heterodimers with Aux/IAA proteins may alter their ability to modulate early auxin response genes expression. Promotes flowering, stamen development, floral organ abscission and fruit dehiscence. Functions independently of ethylene and cytokinin response pathways. May act as a repressor of cell division and organ growth. {ECO:0000269|PubMed:12036261, ECO:0000269|PubMed:15960614, ECO:0000269|PubMed:16176952, ECO:0000269|PubMed:16339187}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00574 | DAP | Transfer from AT5G62000 | Download |
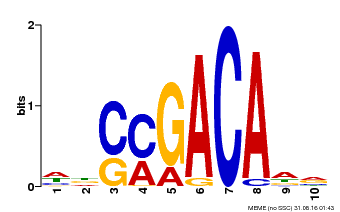
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Lj4g3v2881950.2 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_007159966.1 | 0.0 | hypothetical protein PHAVU_002G282200g | ||||
| Swissprot | Q94JM3 | 0.0 | ARFB_ARATH; Auxin response factor 2 | ||||
| TrEMBL | V7CRR6 | 0.0 | V7CRR6_PHAVU; Auxin response factor | ||||
| STRING | XP_007159966.1 | 0.0 | (Phaseolus vulgaris) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF3281 | 33 | 66 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G62000.4 | 0.0 | auxin response factor 2 | ||||



