 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Lj5g3v1999250.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Loteae; Lotus
|
||||||||
| Family | Nin-like | ||||||||
| Protein Properties | Length: 684aa MW: 74603.8 Da PI: 5.9705 | ||||||||
| Description | Nin-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | RWP-RK | 91.2 | 8.1e-29 | 294 | 344 | 2 | 52 |
RWP-RK 2 ekeisledlskyFslpikdAAkeLgvclTvLKriCRqyGIkRWPhRkiksl 52
ek+isle+l++yF++++kdAAk+Lgvc+T++KriCRq+GI+RWP+Rki+++
Lj5g3v1999250.1 294 EKSISLEVLQRYFAGSLKDAAKSLGVCPTTMKRICRQHGISRWPSRKINKV 344
799**********************************************97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51519 | 17.677 | 283 | 364 | IPR003035 | RWP-RK domain |
| Pfam | PF02042 | 1.1E-25 | 297 | 344 | IPR003035 | RWP-RK domain |
| SuperFamily | SSF54277 | 5.23E-23 | 587 | 674 | No hit | No description |
| Gene3D | G3DSA:3.10.20.240 | 5.2E-26 | 589 | 670 | No hit | No description |
| Pfam | PF00564 | 7.4E-19 | 589 | 670 | IPR000270 | PB1 domain |
| PROSITE profile | PS51745 | 25.247 | 589 | 671 | IPR000270 | PB1 domain |
| SMART | SM00666 | 1.4E-25 | 589 | 671 | IPR000270 | PB1 domain |
| CDD | cd06407 | 6.80E-39 | 590 | 670 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009414 | Biological Process | response to water deprivation | ||||
| GO:0010118 | Biological Process | stomatal movement | ||||
| GO:0010167 | Biological Process | response to nitrate | ||||
| GO:0042128 | Biological Process | nitrate assimilation | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 684 aa Download sequence Send to blast |
MLLNQICNEG RQNALSEILE ILTVVCETHN LPLAQTWVPC RHRSVLAHGG GLKKICSSFD 60 GSCMGQVCMS ITDVASYIID PHLWGFREAC VEHHLQQGQG VAGRAFSSHS MCFCPNITQF 120 CKTDYPLVHY ALMFGLTSCF AICLRSSHTG DDDYVLEFFL PPRITHFSEQ KTLVGSILAT 180 MKQHFQSLKI AAGVELEDGG SVEIIEATNE RVTLRLESIP IAQSVQSPPG PGGSPNMVEE 240 VPKDPSEQQT MMNCNDMNEG GAHGDNAGGN IDQMSSLETK TMKKPSERKR GKTEKSISLE 300 VLQRYFAGSL KDAAKSLGVC PTTMKRICRQ HGISRWPSRK INKVNRSLSK LKRVIESVQG 360 AEGAFGLNSL STSPLPVVGS FPEPSTPNKF SQQASMRPLE SQMKENEIDA SKILEIEDPL 420 LGGMTQNLEK VFNDKGPRRI RTGSGSSEDS TNPTSHGSCY GSPPIESSHV KDISITSNNE 480 RCVVPRGSPE SSPLQPINTL NSPSPIPIPD IVLTELQEPF GGMLIEDAGS SKDLRNLCPS 540 AAEAILEDQV PEACPTNNPP GCSSLAPKRP VDTLGKTVTP FAAKKEMKTV TIKATYREDI 600 IRFRVSLTCG IVELKEEVSK RLKLEVGTFD IKYLDDDQEW VLIACDADLQ ECIDVSRSSA 660 SHIIRVLVHD ITSNLGSSCE SSGE |
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | TISSUE SPECIFICITY: Expressed in roots, stems, leaves, flowers and siliques. Detected in root hairs, emerging secondary roots, vascular tissues, leaf parenchyma cells and stomata. {ECO:0000269|PubMed:18826430}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor involved in regulation of nitrate assimilation and in transduction of the nitrate signal. {ECO:0000269|PubMed:18826430}. | |||||
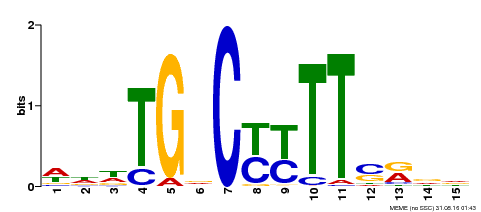
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00449 | DAP | Transfer from AT4G24020 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Lj5g3v1999250.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Not regulated by the N source or by nitrate. {ECO:0000269|PubMed:18826430}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_027363207.1 | 0.0 | protein NLP6-like isoform X1 | ||||
| Swissprot | Q84TH9 | 0.0 | NLP7_ARATH; Protein NLP7 | ||||
| TrEMBL | A0A2Z5V8A6 | 0.0 | A0A2Z5V8A6_LOTJA; Nodule inception-like protein | ||||
| STRING | GLYMA10G37860.2 | 0.0 | (Glycine max) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G24020.1 | 0.0 | NIN like protein 7 | ||||



