 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Lj6g3v0408380.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Loteae; Lotus
|
||||||||
| Family | M-type_MADS | ||||||||
| Protein Properties | Length: 241aa MW: 27506.9 Da PI: 10.2994 | ||||||||
| Description | M-type_MADS family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SRF-TF | 54.8 | 1.2e-17 | 10 | 50 | 2 | 42 |
---SHHHHHHHHHHHHHHHHHHHHHHHHHHT-EEEEEEE-T CS
SRF-TF 2 rienksnrqvtfskRrngilKKAeELSvLCdaevaviifss 42
+i n+s r+ tf kR++g++KK ELS+LC+++++ i++s+
Lj6g3v0408380.1 10 FITNDSARKATFKKRKKGLMKKVAELSTLCGIDACAIVYSP 50
89**************************************8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00432 | 4.2E-25 | 1 | 60 | IPR002100 | Transcription factor, MADS-box |
| PROSITE profile | PS50066 | 16.792 | 1 | 49 | IPR002100 | Transcription factor, MADS-box |
| CDD | cd00266 | 2.66E-23 | 2 | 85 | No hit | No description |
| SuperFamily | SSF55455 | 7.06E-25 | 2 | 95 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 5.7E-8 | 3 | 23 | IPR002100 | Transcription factor, MADS-box |
| Pfam | PF00319 | 1.2E-18 | 10 | 51 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 5.7E-8 | 23 | 38 | IPR002100 | Transcription factor, MADS-box |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0043078 | Cellular Component | polar nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 241 aa Download sequence Send to blast |
MTRKKVKLAF ITNDSARKAT FKKRKKGLMK KVAELSTLCG IDACAIVYSP YDPQPEVWPS 60 PLGVQRVLAK FRRMPELEQS KKMVNQESFL RQRIQKAKEQ LKKQRKDNRE KEITQLMFQC 120 LSAGKILHNI NMVDLNDLAW LIDQNLKDIN RRIEVLTKNA QSQAQAQMVA AAAPPVVTDI 180 VAKIEERSPE GNHGQGLDMN IDVMQKQHWF MNLMNSGGGD EALPLGDVNH QNGFWPNPFF 240 H |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 2 | 24 | RKKVKLAFITNDSARKATFKKRK |
| 2 | 21 | 25 | KKRKK |
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | TISSUE SPECIFICITY: Expressed in the central cell of the female gametophyte and in early endosperm. Also detected in ovaries, young siliques, roots, leaves, stems, young flowers and anthers. {ECO:0000269|PubMed:16798889}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor. Controls central cell differentiation during female gametophyte development. Required for the expression of DEMETER and DD46, but not for the expression of FIS2 (PubMed:16798889). Probable transcription factor that may function in the maintenance of the proper function of the central cell in pollen tube attraction (Probable). {ECO:0000269|PubMed:16798889, ECO:0000305|PubMed:26462908}. | |||||
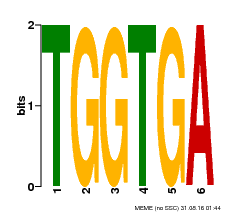
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00553 | DAP | Transfer from AT5G48670 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Lj6g3v0408380.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | - | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AP009689 | 1e-160 | AP009689.1 Lotus japonicus genomic DNA, clone: LjT40B16, TM1372, complete sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_007163366.1 | 1e-140 | hypothetical protein PHAVU_001G228600g | ||||
| Swissprot | Q9FJK3 | 6e-68 | AGL80_ARATH; Agamous-like MADS-box protein AGL80 | ||||
| TrEMBL | V7D1F9 | 1e-138 | V7D1F9_PHAVU; Uncharacterized protein | ||||
| STRING | XP_007163366.1 | 1e-139 | (Phaseolus vulgaris) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF452 | 33 | 154 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G48670.1 | 2e-70 | AGAMOUS-like 80 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



