 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Lus10000492 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Malpighiales; Linaceae; Linum
|
||||||||
| Family | HSF | ||||||||
| Protein Properties | Length: 375aa MW: 42235.5 Da PI: 5.3829 | ||||||||
| Description | HSF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HSF_DNA-bind | 106.2 | 2.7e-33 | 43 | 133 | 2 | 101 |
HHHHHHHHHCTGGGTTTSEESSSSSEEEES-HHHHHHHTHHHHSTT--HHHHHHHHHHTTEEE---SSBTTTTXTTSEEEEESXXXXXXXXXXXXXX CS
HSF_DNA-bind 2 FlkklyeiledeelkeliswsengnsfvvldeeefakkvLpkyFkhsnfaSFvRQLnmYgFkkvkdeekkskskekiweFkhksFkkgkkellekik 98
Fl+k++e+++d +++++sws+ g+sfvv+d+++f++++Lp++Fkh+nf+SFvRQLn+YgF+k++ e+ weF++++F +g+k ll++i+
Lus10000492 43 FLTKTFEMVDDLFTNHVVSWSRGGTSFVVWDPHSFSTNLLPRFFKHNNFSSFVRQLNTYGFRKIDPER---------WEFANEEFLRGHKYLLKNIR 130
9****************************************************************999.........******************** PP
XXX CS
HSF_DNA-bind 99 rkk 101
r+k
Lus10000492 131 RRK 133
*98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.10 | 1.7E-36 | 39 | 132 | IPR011991 | Winged helix-turn-helix DNA-binding domain |
| SuperFamily | SSF46785 | 6.12E-33 | 39 | 132 | IPR011991 | Winged helix-turn-helix DNA-binding domain |
| SMART | SM00415 | 8.1E-57 | 39 | 132 | IPR000232 | Heat shock factor (HSF)-type, DNA-binding |
| Pfam | PF00447 | 1.4E-29 | 43 | 132 | IPR000232 | Heat shock factor (HSF)-type, DNA-binding |
| PRINTS | PR00056 | 3.8E-18 | 43 | 66 | IPR000232 | Heat shock factor (HSF)-type, DNA-binding |
| PRINTS | PR00056 | 3.8E-18 | 81 | 93 | IPR000232 | Heat shock factor (HSF)-type, DNA-binding |
| PROSITE pattern | PS00434 | 0 | 82 | 106 | IPR000232 | Heat shock factor (HSF)-type, DNA-binding |
| PRINTS | PR00056 | 3.8E-18 | 94 | 106 | IPR000232 | Heat shock factor (HSF)-type, DNA-binding |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 375 aa Download sequence Send to blast |
MNNFFTVKEE YPGDTPAMTT AMMTVVPPPQ PMEGLHDAGP PPFLTKTFEM VDDLFTNHVV 60 SWSRGGTSFV VWDPHSFSTN LLPRFFKHNN FSSFVRQLNT YGFRKIDPER WEFANEEFLR 120 GHKYLLKNIR RRKAPLPPPP SLSPSPSPSQ SFYATIPNQA PGTSAVELGQ FGLDGELGRL 180 KRDKQVLTME LVKLRQQQQS TKAYVTELEQ RLQGTESKQQ QMMRFLARAV QNPAFLHQLA 240 QRKEKRKELE EALSKKRRRP IDRGRGKNLV GESSNWRQVI KPEPVKYGIG SPFGLTELEA 300 LALEMQGMGK GVRLEEADGE EEDGEELEAT SDGELDDGFW EELLNESSGG GGEEQEDVNT 360 LAEQLGYLSS SSPK* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5d5u_B | 2e-22 | 38 | 132 | 21 | 129 | Heat shock factor protein 1 |
| 5d5v_B | 2e-22 | 38 | 132 | 21 | 129 | Heat shock factor protein 1 |
| 5d5v_D | 2e-22 | 38 | 132 | 21 | 129 | Heat shock factor protein 1 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 244 | 258 | KRKELEEALSKKRRR |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional activator that specifically binds DNA sequence 5'-AGAAnnTTCT-3' known as heat shock promoter elements (HSE). | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00371 | DAP | Transfer from AT3G22830 | Download |
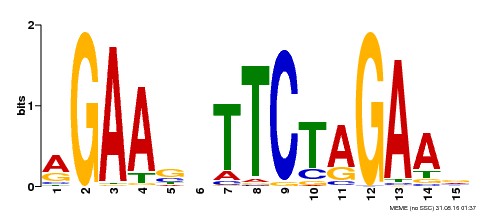
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Lus10000492 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_022759739.1 | 1e-156 | heat stress transcription factor A-7a-like | ||||
| Swissprot | Q9LUH8 | 1e-123 | HFA6B_ARATH; Heat stress transcription factor A-6b | ||||
| TrEMBL | A0A1U8NS09 | 1e-153 | A0A1U8NS09_GOSHI; heat stress transcription factor A-7a-like | ||||
| STRING | Lus10000492 | 0.0 | (Linum usitatissimum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF1100 | 34 | 105 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G22830.1 | 9e-98 | heat shock transcription factor A6B | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Lus10000492 |



