 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Lus10014578 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Malpighiales; Linaceae; Linum
|
||||||||
| Family | M-type_MADS | ||||||||
| Protein Properties | Length: 262aa MW: 28865.2 Da PI: 9.6187 | ||||||||
| Description | M-type_MADS family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SRF-TF | 57.1 | 2.4e-18 | 11 | 52 | 3 | 44 |
--SHHHHHHHHHHHHHHHHHHHHHHHHHHT-EEEEEEE-TTS CS
SRF-TF 3 ienksnrqvtfskRrngilKKAeELSvLCdaevaviifsstg 44
i n+s r+ t+ kR++g++KK +ELS+LCd++++ i++s+ +
Lus10014578 11 IANDSARKATYKKRKKGLMKKVSELSTLCDIQACAIVYSPYD 52
89*************************************965 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF55455 | 1.31E-25 | 1 | 86 | IPR002100 | Transcription factor, MADS-box |
| PROSITE profile | PS50066 | 17.1 | 1 | 49 | IPR002100 | Transcription factor, MADS-box |
| SMART | SM00432 | 3.6E-25 | 1 | 60 | IPR002100 | Transcription factor, MADS-box |
| CDD | cd00266 | 7.81E-21 | 2 | 86 | No hit | No description |
| PRINTS | PR00404 | 6.3E-10 | 3 | 23 | IPR002100 | Transcription factor, MADS-box |
| Pfam | PF00319 | 3.5E-18 | 11 | 55 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 6.3E-10 | 23 | 38 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 6.3E-10 | 38 | 59 | IPR002100 | Transcription factor, MADS-box |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0043078 | Cellular Component | polar nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 262 aa Download sequence Send to blast |
MTRKKVKLAY IANDSARKAT YKKRKKGLMK KVSELSTLCD IQACAIVYSP YDTQPETWPP 60 TAAGVHQVLS RFRKVPQMEQ CKKMVNQEKF LEERIKKAAE QLRKQKRDND EKMVQNALFE 120 CLTGRISLQS LTLGDLDVIA RAVDQQIKEV DRRLAEIARK GGRGGGGGAG GSAATNGVNN 180 LSLEGSLQMM TPANGGGMVG AAKEEFVENV NHENLQRQLW FMEMMNSNAP APLTAVEQQQ 240 MGFMGGEMMM NNVAADDVVH N* |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 2 | 24 | RKKVKLAYIANDSARKATYKKRK |
| 2 | 21 | 25 | KKRKK |
| 3 | 160 | 167 | GGRGGGGG |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00553 | DAP | Transfer from AT5G48670 | Download |
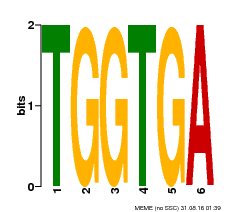
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Lus10014578 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | - | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| STRING | Lus10014578 | 0.0 | (Linum usitatissimum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF452 | 33 | 154 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G48670.1 | 1e-53 | AGAMOUS-like 80 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Lus10014578 |



