 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Lus10024140 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Malpighiales; Linaceae; Linum
|
||||||||
| Family | bZIP | ||||||||
| Protein Properties | Length: 333aa MW: 36885.5 Da PI: 8.4575 | ||||||||
| Description | bZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | bZIP_1 | 30.6 | 7.3e-10 | 45 | 86 | 4 | 45 |
XCHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHH CS
bZIP_1 4 lkrerrkqkNReAArrsRqRKkaeieeLeekvkeLeaeNkaL 45
k rr+++NReAAr+sR+RKka++++Le +L++ ++L
Lus10024140 45 QKTLRRLAQNREAARKSRLRKKAYVQQLESSRLKLTQLEQEL 86
5888*************************9777777666555 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00338 | 5.8E-8 | 42 | 105 | IPR004827 | Basic-leucine zipper domain |
| PROSITE profile | PS50217 | 9.415 | 44 | 88 | IPR004827 | Basic-leucine zipper domain |
| SuperFamily | SSF57959 | 4.99E-7 | 46 | 89 | No hit | No description |
| Pfam | PF00170 | 5.7E-7 | 46 | 86 | IPR004827 | Basic-leucine zipper domain |
| Gene3D | G3DSA:1.20.5.170 | 1.0E-7 | 47 | 91 | No hit | No description |
| PROSITE pattern | PS00036 | 0 | 49 | 64 | IPR004827 | Basic-leucine zipper domain |
| Pfam | PF14144 | 2.1E-33 | 128 | 203 | IPR025422 | Transcription factor TGA like domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009410 | Biological Process | response to xenobiotic stimulus | ||||
| GO:0009627 | Biological Process | systemic acquired resistance | ||||
| GO:0045892 | Biological Process | negative regulation of transcription, DNA-templated | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005737 | Cellular Component | cytoplasm | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 333 aa Download sequence Send to blast |
MADTSPRTDS GDADSDEQSP GFDRGLSGVV AASESSDRSK DKMDQKTLRR LAQNREAARK 60 SRLRKKAYVQ QLESSRLKLT QLEQELQRAR QQGIFISSSG DQAHSMGGNG AMAFDVEYAR 120 WLEEQNKQIN ELRAAVNSHA GDTELRIIID GIMAHYDEIF RLKSNAAKAD VFHLLSGMWK 180 TPAERCFLWL GGFRSSELLK LLVNQLEPLT EQQLVGLTNL QQSSQQAEDA LSQGMEALQQ 240 SLAETLSSGT QPGSSGTSGN VANYMGQMAM AMGKLGTLEG FIRQADNLRQ QTLQQMHRIL 300 TTRQSARALL AIHDYLSRLR ALSSLWLARP RE* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional activator that binds specifically to the DNA sequence 5'-TGACG-3'. Recognizes ocs elements like the as-1 motif of the cauliflower mosaic virus 35S promoter. Binding to the as-1-like cis elements mediate auxin- and salicylic acid-inducible transcription. Binds to the hexamer motif 5'-ACGTCA-3' of histone gene promoters. | |||||
| UniProt | Transcriptional activator that binds specifically to the DNA sequence 5'-TGACG-3'. Recognizes ocs elements like the as-1 motif of the cauliflower mosaic virus 35S promoter. Binding to the as-1-like cis elements mediate auxin- and salicylic acid-inducible transcription. May be involved in the induction of the systemic acquired resistance (SAR) via its interaction with NPR1. Could also bind to the Hex-motif (5'-TGACGTGG-3') another cis-acting element found in plant histone promoters (By similarity). {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00492 | DAP | Transfer from AT5G06950 | Download |
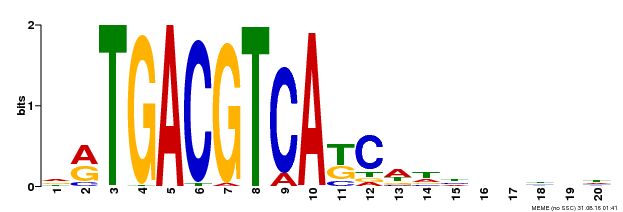
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Lus10024140 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_021649403.1 | 0.0 | transcription factor TGA6-like, partial | ||||
| Refseq | XP_021649414.1 | 0.0 | transcription factor TGA6-like | ||||
| Refseq | XP_021649415.1 | 0.0 | transcription factor TGA6-like | ||||
| Refseq | XP_021649416.1 | 0.0 | transcription factor TGA6-like | ||||
| Refseq | XP_027354195.1 | 0.0 | transcription factor TGA2.2-like isoform X2 | ||||
| Refseq | XP_027354196.1 | 0.0 | transcription factor TGA2.2-like isoform X2 | ||||
| Swissprot | Q39140 | 0.0 | TGA6_ARATH; Transcription factor TGA6 | ||||
| Swissprot | Q41558 | 0.0 | HBP1C_WHEAT; Transcription factor HBP-1b(c1) (Fragment) | ||||
| TrEMBL | B9RG75 | 0.0 | B9RG75_RICCO; Transcription factor HBP-1b(C1), putative | ||||
| STRING | Lus10024140 | 0.0 | (Linum usitatissimum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF1470 | 34 | 94 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G12250.2 | 1e-171 | TGACG motif-binding factor 6 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Lus10024140 |



