 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Lus10026684 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Malpighiales; Linaceae; Linum
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 323aa MW: 35717.6 Da PI: 5.7713 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 51.7 | 1.6e-16 | 63 | 112 | 2 | 55 |
HHHHHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 2 rrahnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
r +h+ E+rRR++iN++f+ Lrel+P++ ++K + a+ L +++eY+++Lq
Lus10026684 63 RSKHSVTEQRRRSKINERFQILRELIPQC----DQKRDTASFLFEVIEYVRYLQ 112
889*************************9....9*******************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF47459 | 1.06E-18 | 59 | 132 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene3D | G3DSA:4.10.280.10 | 5.4E-22 | 61 | 129 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS50888 | 16.228 | 61 | 111 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 2.98E-15 | 63 | 116 | No hit | No description |
| Pfam | PF00010 | 5.7E-14 | 63 | 112 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 1.3E-11 | 67 | 117 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006351 | Biological Process | transcription, DNA-templated | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 323 aa Download sequence Send to blast |
MRGKGNQNHH QEEEEYDEEE LVVGSRKDGG APSSNVSATT SNSANNTNSH RDGKNSDKAS 60 AVRSKHSVTE QRRRSKINER FQILRELIPQ CDQKRDTASF LFEVIEYVRY LQEKVQKYEG 120 PYQGWPADPT KLMPWRNSHW QVQNLVGHPQ AMSNGSGMAP VFPAKLDENG ISVTTSSMME 180 PDSLRDAAMM QQADLLNKDG YPDLLRHFQT AALPAVRSDG VDMHHPLQQP LSDPQSAECP 240 MTAEELATEG GTINISSAYS EGLLNALTQA LQTAGVDLSQ ANISLQIDLG KRAKDNQNTL 300 PSNQTPIHPI DGTDHVDKRM KM* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Positive brassinosteroid-signaling protein. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00219 | DAP | Transfer from AT1G69010 | Download |
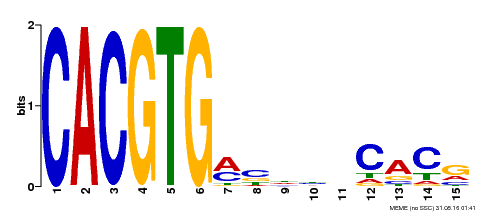
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Lus10026684 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Repressed by heat treatment. {ECO:0000269|PubMed:12679534}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_002314914.2 | 1e-113 | transcription factor BIM2 | ||||
| Refseq | XP_024465346.1 | 1e-113 | transcription factor BIM2 | ||||
| Swissprot | Q9CAA4 | 6e-91 | BIM2_ARATH; Transcription factor BIM2 | ||||
| TrEMBL | B9HWU2 | 1e-112 | B9HWU2_POPTR; Uncharacterized protein | ||||
| STRING | Lus10026684 | 0.0 | (Linum usitatissimum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF1730 | 33 | 82 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G69010.1 | 7e-79 | BES1-interacting Myc-like protein 2 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Lus10026684 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



