 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Lus10028513 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Malpighiales; Linaceae; Linum
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 298aa MW: 33968.1 Da PI: 9.764 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 51.1 | 3.2e-16 | 14 | 61 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g+WT+eEd ll ++++ +G g+W+ ++ + g++R++k+c++rw++yl
Lus10028513 14 KGAWTEEEDRLLRKCIASFGEGRWHQVPLRAGLNRCRKSCRLRWLNYL 61
79********************************************97 PP
| |||||||
| 2 | Myb_DNA-binding | 48.2 | 2.5e-15 | 67 | 111 | 1 | 47 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
rg ++++E +l+ +++k+lG++ W++Ia +++ gRt++++k++w+++
Lus10028513 67 RGEFSEDEVDLITRLHKLLGNR-WSLIAGRLP-GRTANDVKNYWNTH 111
899*******************.*********.************97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.60 | 6.1E-22 | 9 | 64 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 25.528 | 9 | 65 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 1.04E-27 | 11 | 108 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 4.6E-15 | 13 | 63 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 1.8E-15 | 14 | 61 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 1.33E-10 | 16 | 61 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 5.9E-23 | 65 | 115 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 2.1E-15 | 66 | 114 | IPR001005 | SANT/Myb domain |
| PROSITE profile | PS51294 | 18.835 | 66 | 116 | IPR017930 | Myb domain |
| Pfam | PF00249 | 7.0E-14 | 67 | 111 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 3.79E-10 | 69 | 111 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009753 | Biological Process | response to jasmonic acid | ||||
| GO:0031540 | Biological Process | regulation of anthocyanin biosynthetic process | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 298 aa Download sequence Send to blast |
MEGEMQSSTV GLRKGAWTEE EDRLLRKCIA SFGEGRWHQV PLRAGLNRCR KSCRLRWLNY 60 LKPNIKRGEF SEDEVDLITR LHKLLGNRWS LIAGRLPGRT ANDVKNYWNT HQKKKTVSFQ 120 CNPSYTNTNK SEQFKKNNNN AKLQITKTNI IKPRPRTFSS KSFFWPGKKI TTKPQNPVLI 180 INKPRNVDDD VSKKRSYHHM RGSGGESTWV DQLERFLNDG GVVNDDDGSP AAGINGGHEV 240 EVINRTGQLI WSDNDEEKNV NMGDIGTTTM FDGQEDHNQS CWSDIDIWSL LGDHRGI* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1gv2_A | 2e-21 | 12 | 116 | 2 | 105 | MYB PROTO-ONCOGENE PROTEIN |
| 1mse_C | 2e-21 | 12 | 116 | 2 | 105 | C-Myb DNA-Binding Domain |
| 1msf_C | 2e-21 | 12 | 116 | 2 | 105 | C-Myb DNA-Binding Domain |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator, when associated with BHLH002/EGL3/MYC146, BHLH012/MYC1, or BHLH042/TT8. {ECO:0000269|PubMed:15361138}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00213 | DAP | Transfer from AT1G66370 | Download |
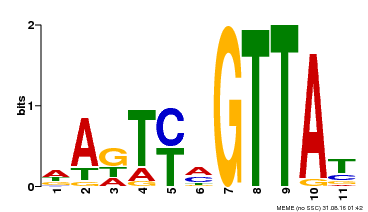
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Lus10028513 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_007033038.1 | 6e-69 | PREDICTED: transcription factor MYB75 isoform X1 | ||||
| Refseq | XP_018816575.1 | 3e-69 | PREDICTED: transcription factor MYB90-like | ||||
| Swissprot | Q9FNV9 | 8e-64 | MY113_ARATH; Transcription factor MYB113 | ||||
| TrEMBL | A0A3G8FWW4 | 1e-68 | A0A3G8FWW4_9SOLA; MYB transcription factor AN2 | ||||
| TrEMBL | A0A3G8FWW5 | 2e-68 | A0A3G8FWW5_PETIN; MYB transcription factor AN2 | ||||
| TrEMBL | A0A3G8FWW7 | 2e-68 | A0A3G8FWW7_9SOLA; MYB transcription factor AN2 | ||||
| TrEMBL | A4GRU5 | 2e-68 | A4GRU5_PETIN; Anthocyanin 2 | ||||
| STRING | Lus10028513 | 0.0 | (Linum usitatissimum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF31 | 34 | 817 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G66370.1 | 4e-66 | myb domain protein 113 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Lus10028513 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



