 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Lus10030935 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Malpighiales; Linaceae; Linum
|
||||||||
| Family | MYB_related | ||||||||
| Protein Properties | Length: 329aa MW: 36294.8 Da PI: 8.345 | ||||||||
| Description | MYB_related family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 40.6 | 5.9e-13 | 5 | 55 | 1 | 46 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHT.....TTS-HHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmg.....kgRtlkqcksrwqk 46
+++WT+eE+e l +++++G+g Wk+I + +R+ ++k++w++
Lus10030935 5 KQKWTAEEEEALRAGIAKHGTGKWKLIQQDPEfnpylSTRSNIDLKDKWRN 55
79***********************************9************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 13.788 | 1 | 61 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 3.37E-16 | 2 | 59 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 1.8E-9 | 4 | 59 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 9.3E-10 | 5 | 55 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 1.3E-14 | 5 | 57 | IPR009057 | Homeodomain-like |
| CDD | cd11660 | 1.82E-20 | 6 | 57 | No hit | No description |
| SMART | SM00526 | 0.0032 | 126 | 193 | IPR005818 | Linker histone H1/H5, domain H15 |
| PROSITE profile | PS51504 | 14.823 | 128 | 198 | IPR005818 | Linker histone H1/H5, domain H15 |
| SuperFamily | SSF46785 | 4.63E-6 | 131 | 204 | IPR011991 | Winged helix-turn-helix DNA-binding domain |
| Gene3D | G3DSA:1.10.10.10 | 2.4E-6 | 131 | 199 | IPR011991 | Winged helix-turn-helix DNA-binding domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006334 | Biological Process | nucleosome assembly | ||||
| GO:0000786 | Cellular Component | nucleosome | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 329 aa Download sequence Send to blast |
MGNPKQKWTA EEEEALRAGI AKHGTGKWKL IQQDPEFNPY LSTRSNIDLK DKWRNIIVTP 60 GEKSRTPKTK VPTADDVVST PLPISQVLDA PPAPIVAAPV PAPVPTPAIL PAVTTDEVAK 120 IAPVDVKICS KYDPMIFEAI TALSCNETSV VDINAIIGFV ERRHELPQNG RKQVTSRLRV 180 LVNREKLEKV QNSYRFKEST PTVVAKAPSP KKQVEQAKGL QRSDSATPIE TLEEAARAAA 240 SIIAEAETKA VRAAEKVHEL EIASILREET ESILLFSQQI YDRCKSESIY SFYHTTYSNF 300 CVFCVRPKVP KSCIFLYVAG MQGETIVI* |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00652 | PBM | Transfer from LOC_Os01g51154 | Download |
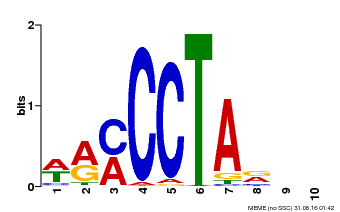
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Lus10030935 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | HQ887128 | 3e-46 | HQ887128.1 Linum usitatissimum clone GMU2AYZ01AG523 microsatellite sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_021650170.1 | 2e-76 | telomere repeat-binding factor 4-like | ||||
| STRING | Lus10030935 | 0.0 | (Linum usitatissimum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF3228 | 29 | 58 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G72740.2 | 5e-44 | MYB_related family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Lus10030935 |



