 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Lus10031321 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Malpighiales; Linaceae; Linum
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 717aa MW: 78657.7 Da PI: 6.3955 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 62.8 | 5e-20 | 39 | 94 | 1 | 56 |
TT--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHHH CS
Homeobox 1 rrkRttftkeqleeLeelFeknrypsaeereeLAkklgLterqVkvWFqNrRakek 56
++k +++t q++ Le++F+++++p++++r L+++lgL+ rq+k+WFqNrR+++k
Lus10031321 39 KKKYHRHTVAQIQKLEAMFKECPHPDEKQRLYLSRELGLSPRQIKFWFQNRRTQMK 94
79999************************************************998 PP
| |||||||
| 2 | START | 171.3 | 6.4e-54 | 231 | 453 | 2 | 206 |
HHHHHHHHHHHHHHHC-TT-EEEE.....EXCCTTEEEEEEESSS.......SCEEEEEEEECCSCHHHHHHHHHCCCGGCT-TT-S....EEEEEEE CS
START 2 laeeaaqelvkkalaeepgWvkss.....esengdevlqkfeeskv......dsgealrasgvvdmvlallveellddkeqWdetla....kaetlev 84
+a a +el+++ +ep+W+ks + +n + + ++f++ + ++ea+r+sgvv+m+ lv ++d + +W e ++ a+t+ev
Lus10031321 231 VAGNAVDELLRLLRTDEPMWMKSLdgggrDVLNLESYQKIFPKPNDhlknpnVRIEASRDSGVVIMNGLALVDMFMDAN-KWMELFPtivsMAKTIEV 327
677899******************99999889999999999776658999999**************************.***999999999****** PP
ECTT......EEEEEEEEXXTTXX-SSX.EEEEEEEEEEE.TTS-EEEEEEEEE-TTS--.-TTSEE-EESSEEEEEEEECTCEEEEEEEE-EE--SS CS
START 85 issg......galqlmvaelqalsplvp.RdfvfvRyirqlgagdwvivdvSvdseqkppesssvvRaellpSgiliepksnghskvtwvehvdlkgr 175
+++g g lql++ elq+lsplvp R+f+++Ry++q ++g+w++v vS d +q+ ++ R+ +lpSg+lie+++ng+skvtwvehv+ ++r
Lus10031321 328 LTPGmigaynGCLQLIYEELQVLSPLVPtREFYILRYCQQIEQGVWAVVNVSYDIPQFASQ----FRSYRLPSGCLIEDMPNGYSKVTWVEHVEIEDR 421
**********************************************************986....9******************************** PP
XX.HHHHHHHHHHHHHHHHHHHHHHTXXXXXX CS
START 176 lp.hwllrslvksglaegaktwvatlqrqcek 206
p h l+r l+ sg+a+ga +w++tlqr ce+
Lus10031321 422 APiHRLYRDLIYSGMAFGAERWLSTLQRMCER 453
******************************97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 1.25E-20 | 25 | 96 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 9.0E-22 | 26 | 98 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50071 | 17.443 | 36 | 96 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 8.1E-20 | 38 | 100 | IPR001356 | Homeobox domain |
| Pfam | PF00046 | 1.3E-17 | 39 | 94 | IPR001356 | Homeobox domain |
| CDD | cd00086 | 1.50E-19 | 39 | 97 | No hit | No description |
| PROSITE pattern | PS00027 | 0 | 71 | 94 | IPR017970 | Homeobox, conserved site |
| PROSITE profile | PS50848 | 51.238 | 221 | 456 | IPR002913 | START domain |
| SuperFamily | SSF55961 | 1.53E-37 | 223 | 454 | No hit | No description |
| CDD | cd08875 | 3.28E-115 | 225 | 452 | No hit | No description |
| SMART | SM00234 | 2.2E-47 | 230 | 453 | IPR002913 | START domain |
| Pfam | PF01852 | 1.6E-46 | 231 | 453 | IPR002913 | START domain |
| Gene3D | G3DSA:3.30.530.20 | 2.7E-7 | 296 | 423 | IPR023393 | START-like domain |
| SuperFamily | SSF55961 | 2.2E-22 | 474 | 707 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009828 | Biological Process | plant-type cell wall loosening | ||||
| GO:0010091 | Biological Process | trichome branching | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0000976 | Molecular Function | transcription regulatory region sequence-specific DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0008289 | Molecular Function | lipid binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 717 aa Download sequence Send to blast |
MEYSNAGGGG ALGGTLGGGG GGAGSGDHDS SSDVQMKKKK KYHRHTVAQI QKLEAMFKEC 60 PHPDEKQRLY LSRELGLSPR QIKFWFQNRR TQMKAQHERA DNCALRAEND KIRCENIAIR 120 EALRNVICPS CGGPPVNEDS YFDEHKLRIE NAQLKEELDR VSSIAGKYIG RPISHLPPVQ 180 PIPVSSLDLS MGNFGGHPLD LDVDLLPQQQ PNLPFQPLGI SSMDKSMMMD VAGNAVDELL 240 RLLRTDEPMW MKSLDGGGRD VLNLESYQKI FPKPNDHLKN PNVRIEASRD SGVVIMNGLA 300 LVDMFMDANK WMELFPTIVS MAKTIEVLTP GMIGAYNGCL QLIYEELQVL SPLVPTREFY 360 ILRYCQQIEQ GVWAVVNVSY DIPQFASQFR SYRLPSGCLI EDMPNGYSKV TWVEHVEIED 420 RAPIHRLYRD LIYSGMAFGA ERWLSTLQRM CERFACLMVS NASTRDLGGV IPSPDGKRSM 480 MKLAQRMVNS FCSSISTSTS HQWTTLSGLD EVGVRVTLRK STDPGQPNGV VLSAATTFWL 540 PVSPQNVFNF FKDEKTRSQW DVLSSGNGVQ EVAHIANGSH PGNCISVLRA FNTSQNNMLI 600 LQESCIDSSG SLVVYCPVEL QAINIAMSGE DPSCIPLLPS GFTICSDGSF GQPSSSADGA 660 STSGKSTGSL ITVAFQILVS SLPSAKLNME SVTTVNNLIG TTVQQIKAAL NCPQSS* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor which acts as positive regulator of drought stress tolerance. Can transactivate CIPK3, NCED3 and ERECTA (PubMed:18451323). Transactivates several cell-wall-loosening protein genes by directly binding to HD motifs in their promoters. These target genes play important roles in coordinating cell-wall extensibility with root development and growth (PubMed:24821957). Transactivates CYP74A/AOS, AOC3, OPR3 and 4CLL5/OPCL1 genes by directly binding to HD motifs in their promoters. These target genes are involved in jasmonate (JA) biosynthesis, and JA signaling affects root architecture by activating auxin signaling, which promotes lateral root formation (PubMed:25752924). Acts as negative regulator of trichome branching (PubMed:16778018, PubMed:24824485). Required for the establishment of giant cell identity on the abaxial side of sepals (PubMed:23095885). May regulate cell differentiation and proliferation during root and shoot meristem development (PubMed:25564655). {ECO:0000269|PubMed:16778018, ECO:0000269|PubMed:18451323, ECO:0000269|PubMed:23095885, ECO:0000269|PubMed:24821957, ECO:0000269|PubMed:24824485, ECO:0000269|PubMed:25564655, ECO:0000269|PubMed:25752924}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00232 | DAP | Transfer from AT1G73360 | Download |
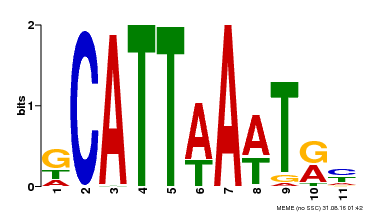
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Lus10031321 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_017973835.1 | 0.0 | PREDICTED: homeobox-leucine zipper protein HDG11 | ||||
| Swissprot | Q9FX31 | 0.0 | HDG11_ARATH; Homeobox-leucine zipper protein HDG11 | ||||
| TrEMBL | A0A2P5B295 | 0.0 | A0A2P5B295_TREOI; Octamer-binding transcription factor | ||||
| STRING | Lus10031321 | 0.0 | (Linum usitatissimum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF1026 | 33 | 98 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G73360.1 | 0.0 | homeodomain GLABROUS 11 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Lus10031321 |



