 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Lus10031817 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Malpighiales; Linaceae; Linum
|
||||||||
| Family | EIL | ||||||||
| Protein Properties | Length: 586aa MW: 65396.3 Da PI: 5.4498 | ||||||||
| Description | EIL family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | EIN3 | 431.4 | 8.8e-132 | 38 | 382 | 1 | 322 |
XXXXXXXXXXXXXXXXXXXXXXX..XXXXX.XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX CS
EIN3 1 eelkkrmwkdqmllkrlkerkkqlledkeaatgakksnksneqarrkkmsraQDgiLkYMlkemevcnaqGfvYgiipekgkpvegasdsLraWWkek 98
+el++rmwkd+++l+r+ker++ + ++ a++++k +++++qa+rkkmsraQDgiLkYMlk mevc+a+GfvYgiipekgkpv+gasd++raWWkek
Lus10031817 38 DELERRMWKDRVKLQRIKERRRISA--QQ-AAEKQKPKHTSDQAKRKKMSRAQDGILKYMLKLMEVCQARGFVYGIIPEKGKPVSGASDNIRAWWKEK 132
79******************99754..44.599***************************************************************** PP
XXXXXXXXXXXXXXXXXXXXXXXXXXX....XX----STTS-HHHHHHHHHHHSSSSSS-TTS--TTT--HHHH---S--HHHHHHT--TT--.---- CS
EIN3 99 vefdrngpaaiskyqaknlilsgesslqtersseshslselqDTtlgSLLsalmqhcdppqrrfplekgvepPWWPtGkelwwgelglskdqgtppyk 196
v+fd+ngpaai+ky+a++l+++++++ ++ ++++ l++lqD+tlgSLLs+lmqhcdppqr++plekgv+pPWWPtG+e+wwg+lgl+ q ppyk
Lus10031817 133 VKFDKNGPAAIAKYEAECLAMNEGDNA--RQGNSQSVLQDLQDATLGSLLSSLMQHCDPPQRKYPLEKGVPPPWWPTGDEDWWGKLGLPGGQT-PPYK 227
********************9988877..789999******************************************************9987.**** PP
-GGG--HHHHHHHHHHHHHHTGGGHHHHHHTTTTSSSSTTT--SHHHHHHHHHHTTTTT-S--XXXXXX.......XXXX..XXXXXXXXXXXXXXXX CS
EIN3 197 kphdlkkawkvsvLtavikhmsptieeirelerqskylqdkmsakesfallsvlnqeekecatvsahss.......slrk..qspkvtlsceqkedve 285
kphdlkk+wkv+vLtavikhmsp+i++ir+++rqsk+lqdkm+akes+++l+vl++ee+ +++ s+++ + ++++ +s+++++dve
Lus10031817 228 KPHDLKKVWKVGVLTAVIKHMSPNIAKIRKHVRQSKCLQDKMTAKESAIWLAVLSREESVIRQPSSDNGtsgitevP--RggRKKQPAVSDDSDYDVE 323
*******************************************************************4444566542..1458888999********* PP
XXXXXX.XXXXXXXXXX.......................XXXXXXXXXXXXXXXXXXXX CS
EIN3 286 gkkeskikhvqavktta.......................gfpvvrkrkkkpsesakvss 322
g ++ + +++++ + + rkrk+ p++++k s+
Lus10031817 324 GVDDGVGSVSSKDSMSNhpmdveppsnlrddsphpvqekeRREKPRKRKR-PRDKSKDSE 382
66666656666666666777799999999988776666544444444443.333333333 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF04873 | 3.7E-126 | 38 | 285 | No hit | No description |
| Gene3D | G3DSA:1.10.3180.10 | 2.5E-71 | 160 | 293 | IPR023278 | Ethylene insensitive 3-like protein, DNA-binding domain |
| SuperFamily | SSF116768 | 1.12E-61 | 166 | 288 | IPR023278 | Ethylene insensitive 3-like protein, DNA-binding domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0042762 | Biological Process | regulation of sulfur metabolic process | ||||
| GO:0071281 | Biological Process | cellular response to iron ion | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 586 aa Download sequence Send to blast |
MGELEEIAAD ICSEIDADEL KCDNLAEKDV SDEEIEPDEL ERRMWKDRVK LQRIKERRRI 60 SAQQAAEKQK PKHTSDQAKR KKMSRAQDGI LKYMLKLMEV CQARGFVYGI IPEKGKPVSG 120 ASDNIRAWWK EKVKFDKNGP AAIAKYEAEC LAMNEGDNAR QGNSQSVLQD LQDATLGSLL 180 SSLMQHCDPP QRKYPLEKGV PPPWWPTGDE DWWGKLGLPG GQTPPYKKPH DLKKVWKVGV 240 LTAVIKHMSP NIAKIRKHVR QSKCLQDKMT AKESAIWLAV LSREESVIRQ PSSDNGTSGI 300 TEVPRGGRKK QPAVSDDSDY DVEGVDDGVG SVSSKDSMSN HPMDVEPPSN LRDDSPHPVQ 360 EKERREKPRK RKRPRDKSKD SEQQPGPSTN DGHFLLEQRA SLPDMDHTHI RALEPQMHAV 420 DLGINTNVTT MPLGEGLEGE PNFNIPEFNY LSDIPPGSGE NICLDGRQQL YPLAQSSDMF 480 HGSSYNFYAP TVNFGPAHGV EQQTHLPMDA SHIRQEGDGF QVSAIPMNEN EIPGGDLQNF 540 AKDIFPSGQD RYLGSPFDSV SLDFGDFSSP CDFDMDKDLL KCYGA* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wij_A | 5e-73 | 165 | 296 | 9 | 140 | ETHYLENE-INSENSITIVE3-like 3 protein |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 366 | 374 | KPRKRKRPR |
| 2 | 367 | 373 | PRKRKRP |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor that may be involved in the ethylene response pathway. {ECO:0000269|PubMed:9215635, ECO:0000269|PubMed:9851977}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00233 | DAP | Transfer from AT1G73730 | Download |
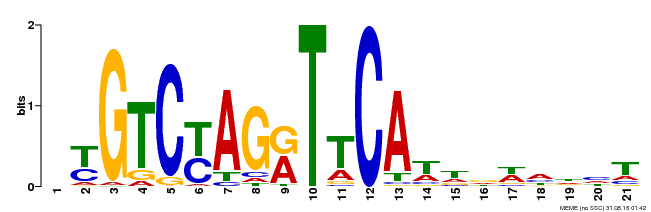
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Lus10031817 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_002299248.2 | 0.0 | ETHYLENE INSENSITIVE 3-like 3 protein | ||||
| Swissprot | O23116 | 0.0 | EIL3_ARATH; ETHYLENE INSENSITIVE 3-like 3 protein | ||||
| TrEMBL | A0A1Q3C887 | 0.0 | A0A1Q3C887_CEPFO; EIN3 domain-containing protein | ||||
| STRING | Lus10031817 | 0.0 | (Linum usitatissimum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF322 | 34 | 200 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G73730.1 | 1e-170 | ETHYLENE-INSENSITIVE3-like 3 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Lus10031817 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



