 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Lus10035504 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Malpighiales; Linaceae; Linum
|
||||||||
| Family | Dof | ||||||||
| Protein Properties | Length: 152aa MW: 16563.3 Da PI: 9.0067 | ||||||||
| Description | Dof family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-Dof | 120.4 | 6.3e-38 | 21 | 84 | 2 | 62 |
zf-Dof 2 kekalkcprCdstntkfCyynnyslsqPryfCkaCrryWtkGGalrnvPvGg...grrknkkss 62
++++l+cprCds+ntkfCyynny+lsqPr+fCk CrryWtkGGalrn+PvGg g+rkn k+s
Lus10035504 21 EQEQLNCPRCDSSNTKFCYYNNYNLSQPRFFCKDCRRYWTKGGALRNIPVGGgggGTRKNAKRS 84
57899*********************************************86222689998776 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF02701 | 7.4E-32 | 23 | 80 | IPR003851 | Zinc finger, Dof-type |
| ProDom | PD007478 | 5.0E-20 | 25 | 83 | IPR003851 | Zinc finger, Dof-type |
| PROSITE profile | PS50884 | 27.41 | 25 | 79 | IPR003851 | Zinc finger, Dof-type |
| PROSITE pattern | PS01361 | 0 | 27 | 63 | IPR003851 | Zinc finger, Dof-type |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0010200 | Biological Process | response to chitin | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 152 aa Download sequence Send to blast |
MQDPTSEFPM IHQQPAPPFP EQEQLNCPRC DSSNTKFCYY NNYNLSQPRF FCKDCRRYWT 60 KGGALRNIPV GGGGGGTRKN AKRSSNSNQK RANPDPTPDS SPSVTRRLVS EPTSPPSSSS 120 VPLGADQDPT RLYGPEPGRD TGLGSVLSQF P* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
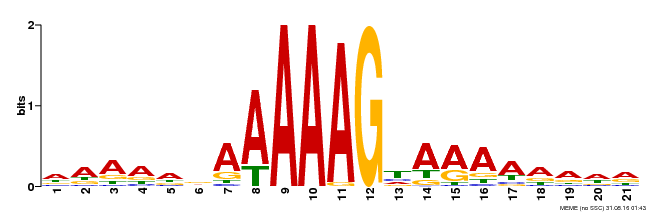
| UniProt | Transcription factor that binds specifically to a 5'-AA[AG]G-3' consensus core sequence. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00199 | DAP | Transfer from AT1G51700 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Lus10035504 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_021645559.1 | 2e-48 | dof zinc finger protein DOF3.1-like | ||||
| Refseq | XP_021645560.1 | 2e-48 | dof zinc finger protein DOF3.1-like | ||||
| Swissprot | O82155 | 3e-39 | DOF17_ARATH; Dof zinc finger protein DOF1.7 | ||||
| TrEMBL | A0A3Q9EDL7 | 7e-46 | A0A3Q9EDL7_POPTO; DOF zinc finger protein 1 | ||||
| STRING | Lus10035504 | 1e-107 | (Linum usitatissimum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF6181 | 33 | 51 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G51700.1 | 1e-29 | DOF zinc finger protein 1 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Lus10035504 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



